Literature Sharing | Chinese cabbage orphan gene BR3 confers bolting resistance to Arabidopsis through the gibberellin pathway
Release time:
2025-07-08
Premature bolting affects yield and quality in Chinese cabbage, highlighting the importance of identifying bolting resistance genes. This study identifies an orphan gene, BR3 (BOLTING RESISTANCE 3), in Arabidopsis thaliana as a positive regulator of bolting resistance. BR3 is expressed during the seedling and flowering stages and localizes to the plasma membrane and nucleus. Overexpression of BR3 (BR3OE) delays bolting and flowering under both long-day and short-day conditions, with increased rosette leaf number and reduced plant height. Key flowering genes are downregulated in BR3OE plants. GA₃ treatment induces early flowering in both BR3OE and wild type (WT) plants, but BR3OE still flowers later than WT. In Chinese cabbage, BR3 shows co-expression with DELLA genes BrRGA1 and BrRGL3, suggesting a regulatory role through the GA pathway. This study offers new insights into bolting resistance mechanisms and provides valuable targets for breeding bolting-resistant Chinese cabbage varieties.
The BR3 gene (BraA07003496) in Chinese cabbage is 347 bp long, located on chromosome A07, and contains two exons and one intron, encoding a 76-amino acid protein. Bioinformatic analyses revealed that BR3 lacks conserved domains, transcription factor activity, kinase function, signal peptides, cleavage sites, and transmembrane regions, indicating it is a novel gene with an unknown function. Structural predictions show it mainly comprises random coils (42.11%).
Expression analysis showed that BR3 is expressed during seedling development, with peak expression at 6 and 8 days after the emergence of the first true leaf. At 4 days post-flowering, BR3 was expressed in multiple organs, with the highest level in flowers. These findings suggest that BR3 may play a direct or indirect role in bolting resistance in Chinese cabbage.
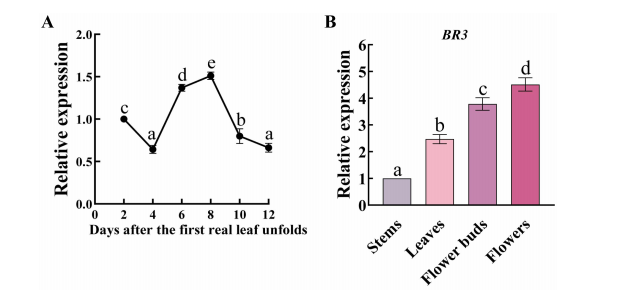
FIGURE 1 Expression patterns of the BR3 gene in Chinese cabbage.
To investigate the spatiotemporal expression of the BR3 gene, GUS staining was performed in transgenic Arabidopsis plants. Strong GUS signals were observed in flower buds, leaves, and roots, indicating BR3 is expressed in these tissues after flowering. Subcellular localization analysis using a 35S::BR3::GFP construct in Nicotiana benthamiana revealed that BR3 localizes to both the nucleus and plasma membrane. These results lay the groundwork for understanding the cellular mechanisms by which BR3 regulates flowering.
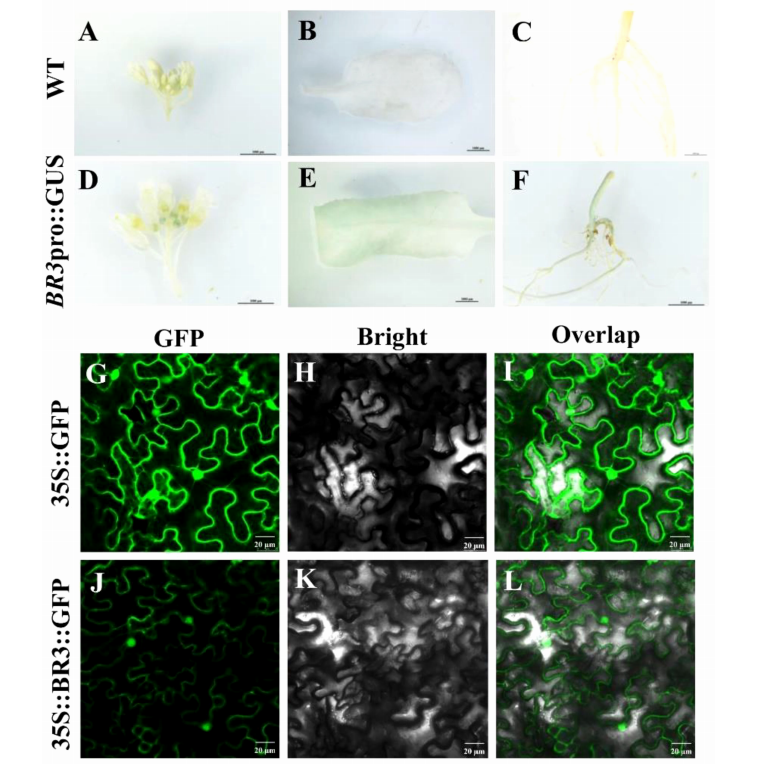
FIGURE 2 Expression analysis of BR3 gene promoter and subcellular localization of BR3.
To assess whether BR3 affects flowering through the photoperiod pathway, flowering time was compared between WT and BR3 overexpression (BR3OE) plants under long-day (LD) and short-day (SD) conditions. Under LD conditions, BR3OE plants showed delayed bolting and flowering by approximately 8.6 and 8.5 days, respectively, compared to WT. They also exhibited reduced plant height, fewer siliques, and more rosette leaves. Another BR3OE line (BR3OE#2) showed a consistent phenotype. qRT-PCR analysis revealed that key flowering genes AtFT, AtSOC1, and AtLFY were significantly downregulated in BR3OE plants. These findings indicate that BR3 delays flowering in Arabidopsis by repressing genes in the photoperiod pathway.
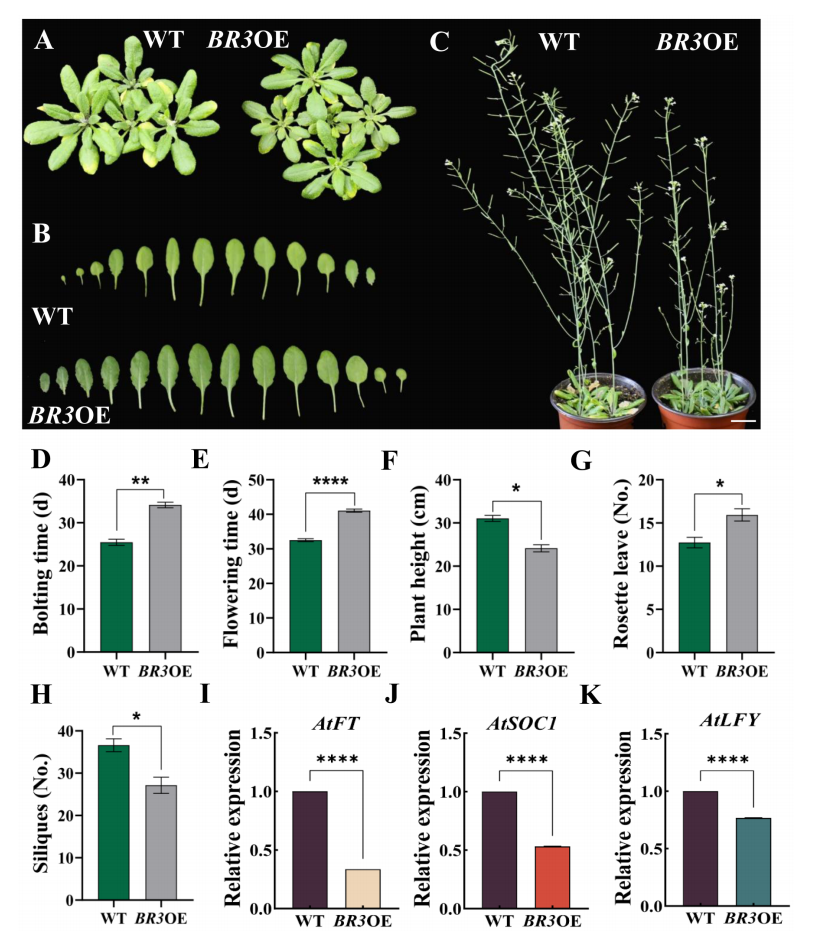
FIGURE 3 Phenotypes of WT and BR3OE under LD conditions and expression of key flowering genes
Under short-day (SD) conditions, BR3 overexpression (BR3OE) plants showed a significant delay in bolting (by 34.87 days) and flowering (by 35.27 days) compared to wild-type (WT) plants. BR3OE plants also had reduced plant height and more rosette leaves, indicating enhanced vegetative growth. Since the late-flowering phenotype occurred under both long-day and short-day conditions, BR3 likely regulates flowering independently of the photoperiod. These findings suggest that BR3 enhances bolting resistance by promoting vegetative growth and delaying the transition to reproductive development.
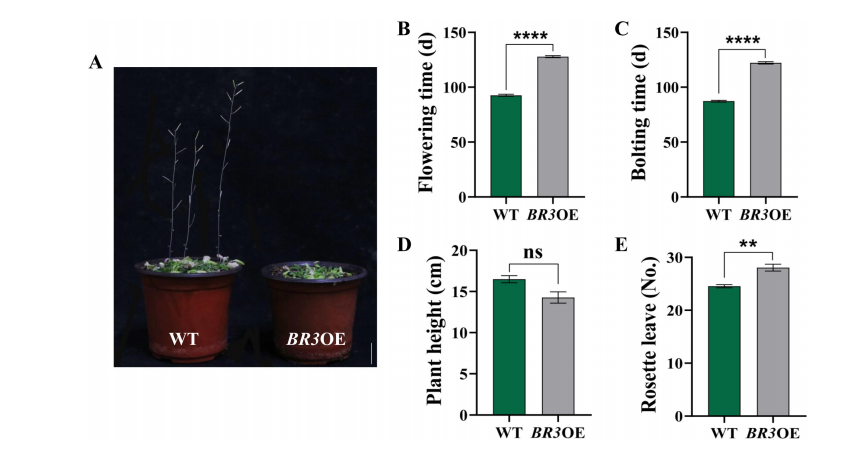
FIGURE 4 Phenotypes of WT and BR3OE plants under SD
Vernalization accelerated bolting and flowering in wild-type (WT) plants by approximately 4.3 days, increased plant height, and reduced the number of rosette leaves. In contrast, BR3 overexpression (BR3OE) plants showed no significant changes in bolting time, flowering time, or rosette leaf number after vernalization. These findings suggest that BR3 delays flowering independently of the vernalization pathway, likely acting through alternative regulatory mechanisms.
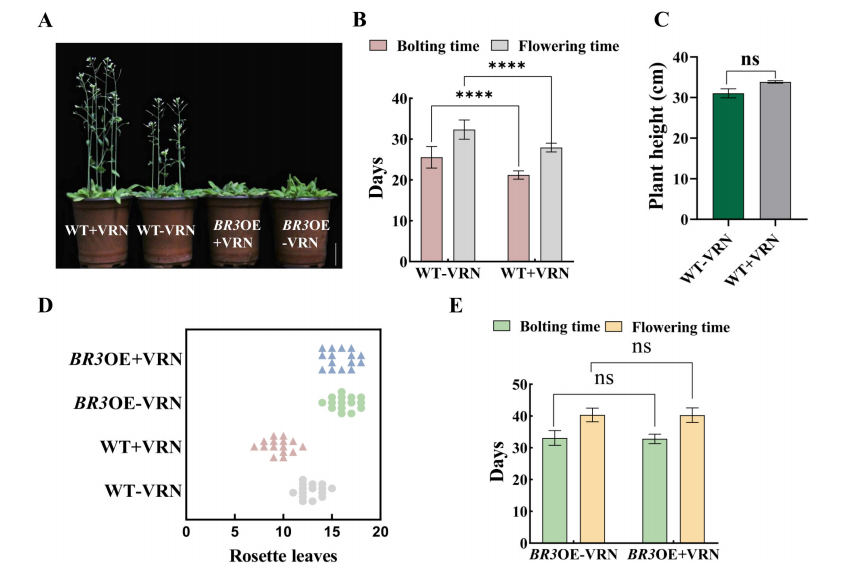
FIGURE 5 Agronomic traits in WT and BR3OE plants after vernalization treatment
Exogenous GA₃ treatment accelerated bolting and flowering in both WT and BR3 overexpression (BR3OE) plants. In WT, bolting and flowering occurred ~4.4 days earlier, plant height increased, and rosette leaf number decreased. In BR3OE plants, flowering was also promoted (~5 days earlier), and rosette leaf number was reduced. Despite GA₃ treatment, BR3OE plants still flowered later than WT, suggesting BR3 delays flowering by modulating GA signaling.
qRT-PCR analysis of five DELLA genes in Brassica rapa showed that BrRGA1 and BrRGL3 expression levels increased in response to repeated GA₃ treatments, mirroring BR3 expression. This indicates that BR3 may influence flowering by upregulating specific DELLA genes in the GA pathway, contributing to bolting resistance.
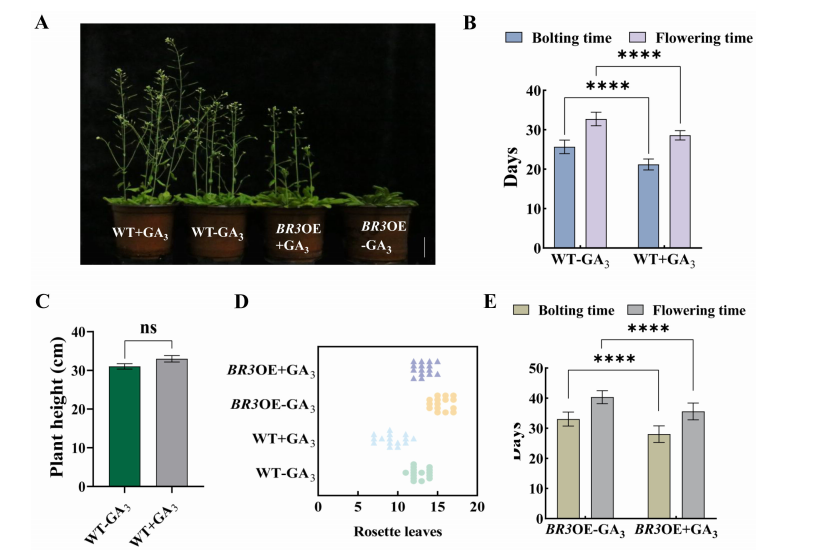
FIGURE 6 Agronomic traits in WT and BR3OE plants treated with GA3
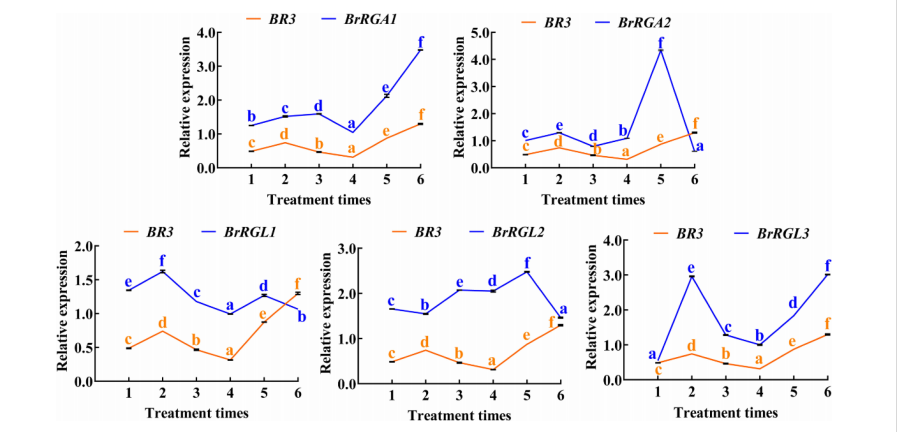
FIGURE 7 Expression analysis of BR3 and DELLA genes in Chinese cabbage after GA3 treatment
This study identified a novel orphan gene (OG), BR3, as a positive regulator of bolting resistance, highlighting the role of OGs in species-specific trait development. BR3 was highly expressed in flower-related tissues and localized to the nucleus and plasma membrane. Overexpression of BR3 (BR3OE) resulted in delayed bolting and downregulation of key flowering genes. Exogenous GA₃ treatment and DELLA gene expression analysis suggest that BR3 regulates flowering time through the gibberellin signaling pathway. These findings offer valuable insights for breeding bolting-resistant Chinese cabbage and lay the groundwork for future research on bolting resistance mechanisms.
Related News
2025-07-10
2025-07-08
2025-07-03
Literature Sharing | Regulation of co-translational mRNA decay by PAP and DXO1 in Arabidopsis
2025-07-01
2025-06-27
2025-06-24
2025-06-20



