Literature Sharing | Regulation of co-translational mRNA decay by PAP and DXO1 in Arabidopsis
Release time:
2025-07-03
This study investigates the regulation of the co-translational mRNA decay (CTRD) pathway in Arabidopsis, a critical mechanism for maintaining mRNA homeostasis. The researchers found that 3ʹ-phosphoadenosine 5ʹ-phosphate (PAP), an inhibitor of exoribonucleases, affects CTRD activity. Specifically, they showed that loss of FRY1 impairs XRN4-dependent CTRD and that exogenous PAP treatment stabilizes CTRD target mRNAs. Additionally, they discovered that another PAP-sensitive exoribonuclease, DXO1, also contributes to CTRD, likely by acting on NAD⁺-capped mRNAs involved in stress responses. These findings reveal new layers of regulation and additional players in the CTRD pathway in plants.

This study explores the role of FRY1 and its downstream metabolite PAP in regulating co-translational mRNA decay (CTRD) in Arabidopsis. PAP, an inhibitor of XRN exoribonucleases, accumulates in fry1 mutants due to loss of FRY1 phosphatase activity. The researchers examined XRN4 association with polysomes (a proxy for CTRD activity) and found reduced XRN4 accumulation in polysome fractions of fry1 mutants. Using 5′P sequencing, they showed that both xrn4 and fry1 mutants exhibited a significant reduction in the characteristic CTRD-associated 5′P signal near stop codons. This reduction was quantified using the Terminational Stalling Index (TSI), which dropped more drastically in fry1 than in xrn4, suggesting stronger impairment of CTRD in fry1. A large overlap of target transcripts was identified between fry1 and xrn4 mutants, indicating that FRY1 acts through XRN4 to regulate CTRD. However, the more severe effect in fry1 suggests that additional PAP-sensitive enzymes besides XRN4 may also contribute to CTRD regulation.
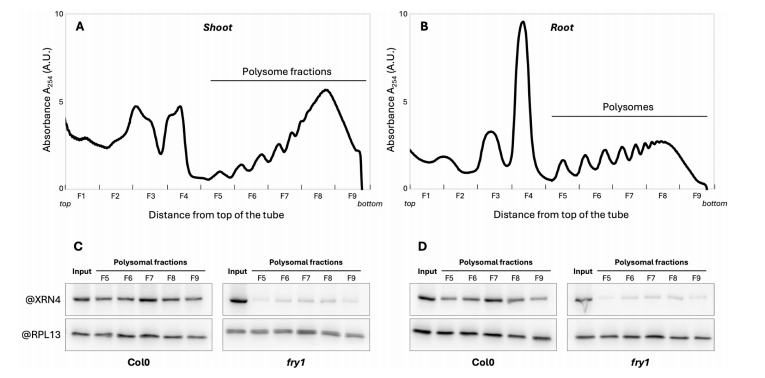
Fig. 1 XRN4 accumulation to polysomes is impaired in fry1mutant. Polysomal extracts prepared from Col0 or fry1 shoot
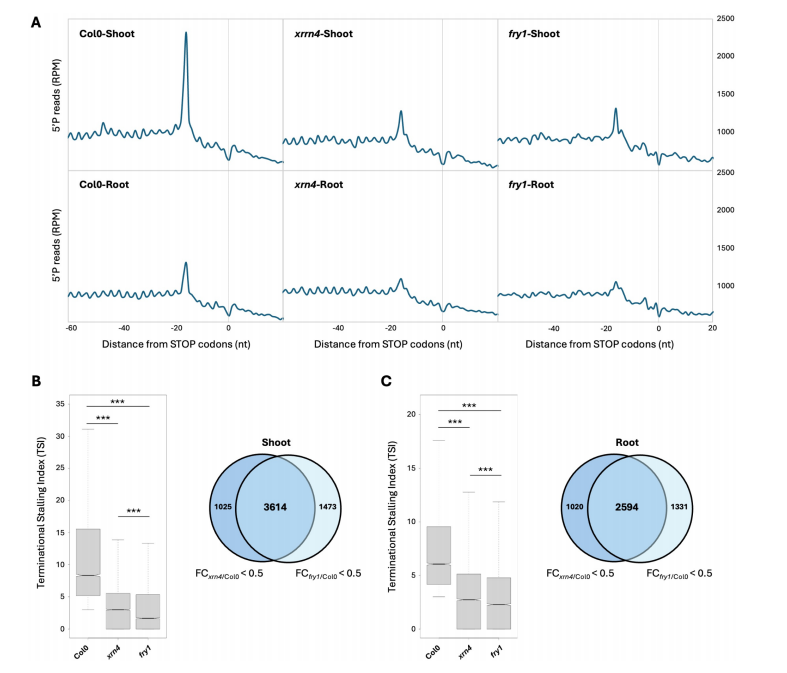
Fig. 2 FRY1 inactivation affects co-translational mRNA decay in both shoot and root
This part of the study examines whether exogenous application of PAP can influence CTRD activity in Arabidopsis. Seedlings were treated with 1 mM PAP (along with ATP to facilitate uptake), and polysome profiling revealed that PAP treatment reduces XRN4 accumulation in polysome fractions in both shoots and roots, without affecting overall XRN4 protein levels. Instead, XRN4 shifted to the free mRNP fraction, indicating impaired association with translating ribosomes. To assess functional consequences, mRNA stability of two known CTRD target transcripts was measured after cordycepin treatment with or without PAP. PAP significantly increased the half-lives of these mRNAs in both shoots and roots, confirming that PAP stabilizes CTRD targets. In contrast, non-CTRD targets were unaffected. These findings demonstrate that PAP treatment can inhibit CTRD activity by interfering with XRN4 function, likely mimicking the effect seen in fry1 mutants.
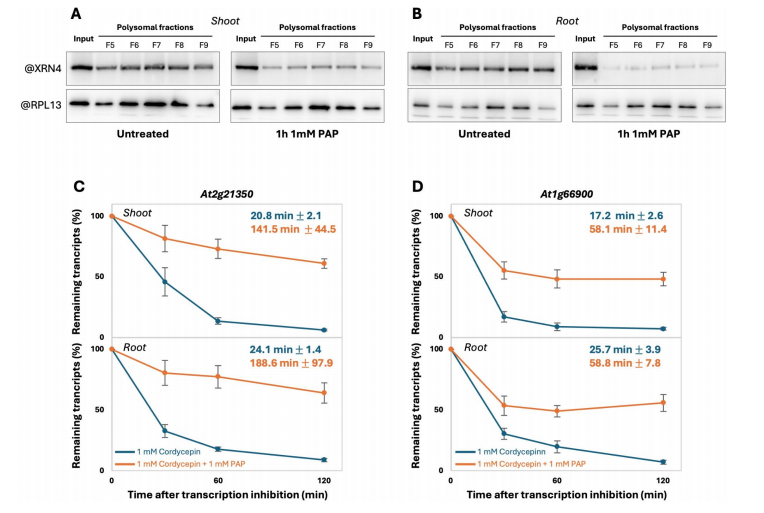
Fig. 3 PAP treatment affects XRN4 accumulation to polysomes and mRNA stability.
This section investigates the potential role of the exoribonuclease DXO1 in the co-translational mRNA decay (CTRD) pathway. Since PAP has been shown to inhibit DXO1 activity in vitro, the researchers examined whether DXO1 is involved in CTRD regulation. Using a DXO1-GFP fusion line, they found that DXO1 associates with ribosomes, with a stronger signal in ribosome-bound fractions—similar to XRN4. This ribosome association was reduced after PAP treatment, suggesting PAP also affects DXO1 localization.
To assess DXO1’s functional role in CTRD, a catalytically inactive mutant (DXO1(E394A/D396A)) was analyzed using 5′P sequencing. Compared to wild-type (Col0), this mutant showed a moderate but significant reduction in CTRD signal near stop codons in both shoot and root, as indicated by metagene and TSI analyses. Although the effect was weaker than in the xrn4 mutant, there was a substantial overlap in CTRD targets between DXO1 and XRN4 (over 85% in shoot and ~74% in root), suggesting that DXO1 contributes to CTRD and shares many targets with XRN4.
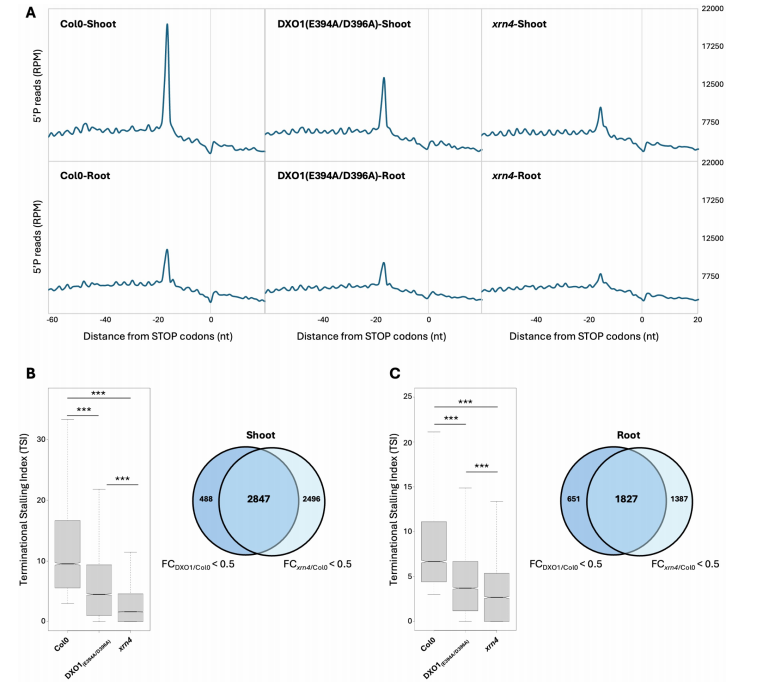
Fig. 4 DXO1 catalytic inactivation affects co-translational mRNA decay in both shoot and root.
This section examines whether NAD⁺-capped mRNAs are subject to co-translational decay (CTRD) and whether DXO1 plays a role in their turnover. By comparing DXO1 CTRD targets with previously identified NAD⁺-capped transcripts (from SPAAC-NAD-seq data), the authors found that a large proportion (83.3%) of NAD⁺-capped mRNAs are CTRD targets in wild-type plants (TSI > 3). In the catalytically inactive DXO1(E394A/D396A) line, TSI values for these transcripts were significantly reduced, indicating impaired decay.
A significant overlap was observed between NAD⁺-capped CTRD targets and transcripts stabilized in the DXO1 mutant, suggesting that DXO1 contributes to their degradation. Gene Ontology analysis of these targets revealed enrichment in stress-related pathways, particularly responses to abscisic acid (ABA), aligning with previous reports that DXO1 regulates ABA-related NAD⁺-capped mRNAs.
Finally, comparison with fry1 mutants showed that PAP-sensitive DXO1 and FRY1 share overlapping targets, although the fry1 mutation had a stronger effect. Altogether, these findings suggest that NAD⁺-capped transcripts can undergo co-translational decay, likely mediated by DXO1, and that this process is sensitive to PAP accumulation.
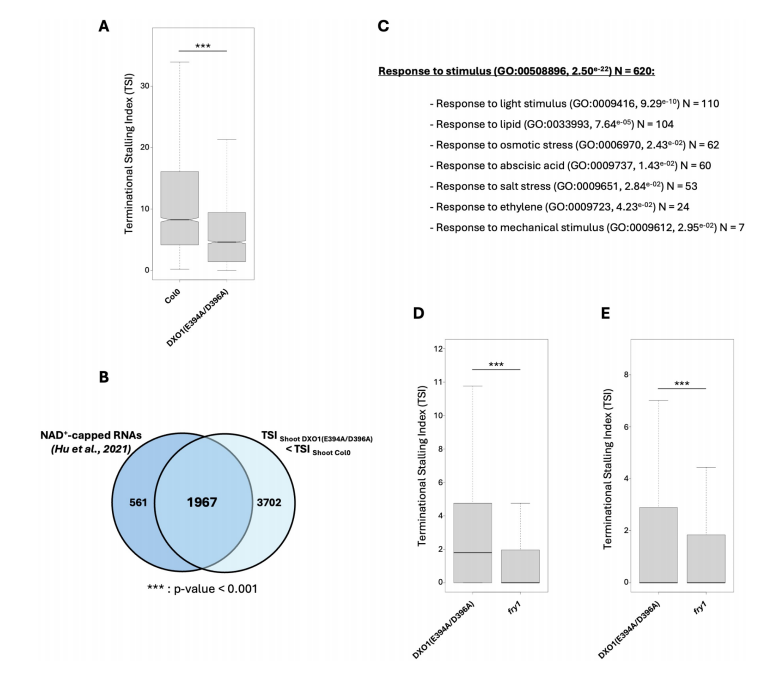
Fig. 5 DXO1 co-translational mRNA decay targets are identified as NAD+-capped RNAs and targeted by FRY1.
This study reveals that the co-translational mRNA decay (CTRD) pathway in Arabidopsis is regulated by PAP and involves the exoribonuclease DXO1, which likely targets NAD⁺-capped mRNAs. These findings uncover a new layer of CTRD regulation and provide a foundation for further exploration of its biological significance in plants.
Related News
2025-07-10
2025-07-08
2025-07-03
Literature Sharing | Regulation of co-translational mRNA decay by PAP and DXO1 in Arabidopsis
2025-07-01
2025-06-27
2025-06-24
2025-06-20



