Literature Sharing | Interaction of PsMYB4 with PsEGL3 inhibits anthocyanin biosynthesis in tree peony yellow flowers
Release time:
2025-07-16
This study explores the molecular mechanism behind yellow flower formation in tree peony, a highly valued ornamental plant in China. Researchers identified two transcription factors, PsMYB4 and PsEGL3, that are highly expressed in a yellow-flowered cultivar. Phylogenetic analysis showed that PsMYB4 is an R2R3-MYB repressor and PsEGL3 belongs to the bHLH IIIf subgroup. Overexpression of these genes in tobacco suppressed anthocyanin biosynthesis, particularly in PsMYB4 lines. Silencing either gene in tree peony petals increased expression of key anthocyanin biosynthetic genes. Interaction between PsMYB4 and PsEGL3 was confirmed through Y2H and BiFC assays, and dual-luciferase assays demonstrated their synergistic repression of anthocyanin-related gene promoters. The results suggest that PsMYB4 and PsEGL3 jointly inhibit anthocyanin production, redirecting metabolic flux toward flavonol synthesis, thereby contributing to yellow pigmentation in tree peony flowers.
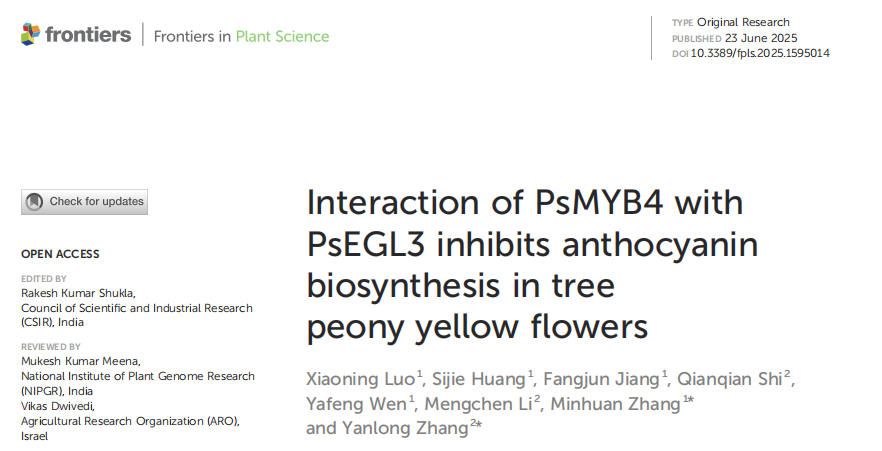
Six tree peony cultivars—‘Baixueta’, ‘Yaohuang’, ‘Huangguan’, ‘High Noon’, ‘Jinge’, and ‘Roufurong’—were grown under optimal light and moisture conditions at the Tree Peony Garden of Northwest A&F University in Shaanxi, China. Petal samples from nine plants per cultivar were collected at five defined developmental stages, with floral spots removed from certain cultivars. Tobacco plants (N. tabacum and N. benthamiana) were grown under controlled conditions (16 h light/8 h dark at 25°C). Petal color parameters (L*, a*, b*, C*, and h) were measured using a color meter, while petals for other analyses were frozen in liquid nitrogen and stored at -80°C.
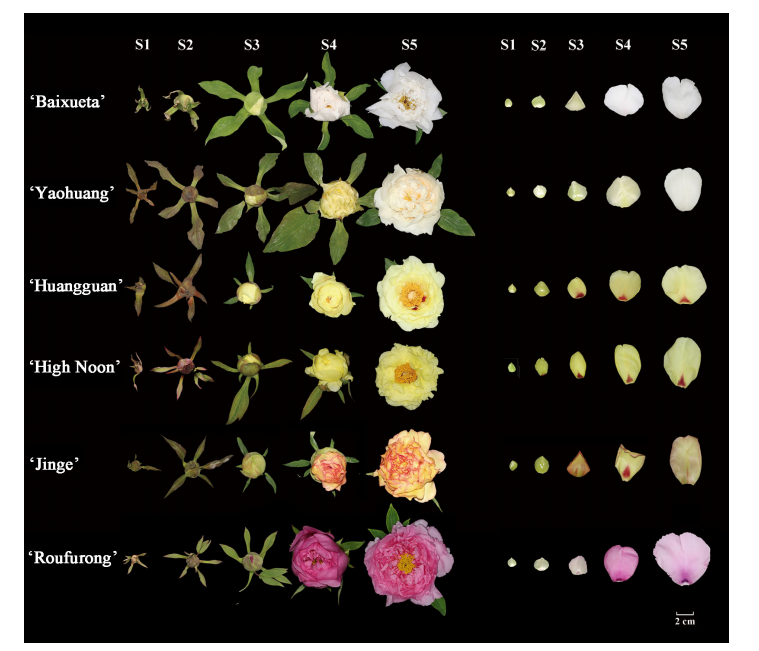
Figure 1. Floral phenotypes of tree peony cultivars at five blooming stages.
This study analyzed flavonoid composition in four tree peony cultivars—‘Baixueta’ (white), ‘Yaohuang’ (yellowish), ‘Huangguan’ (light yellow), and ‘Jinge’ (reddish yellow)—at five floral stages using HPLC, complementing previous research on ‘High Noon’ (yellow) and ‘Roufurong’ (purple-red). Anthocyanins were only detected in ‘Jinge’ and ‘Roufurong’, aligning with their red pigmentation. ‘High Noon’ showed high tetrahydroxychalcone (THC) levels, contributing to its yellow coloration alongside abundant flavones and flavonols like kaempferol (Km), apigenin, chrysoeriol, and isorhamnetin. In contrast, the other four cultivars had lower THC but higher flavone and flavonol levels. Notably, ‘Baixueta’ and ‘Yaohuang’ had lower overall flavonoid levels, possibly explaining their lighter colors. Kaempferol was the dominant flavonol in ‘Baixueta’, ‘Huangguan’, and ‘Jinge’, with consistent trends in the first two. In ‘Jinge’, Km levels peaked mid-development, while anthocyanin levels also peaked at stage S3 before declining, mirroring the fading of red hues in its petals.
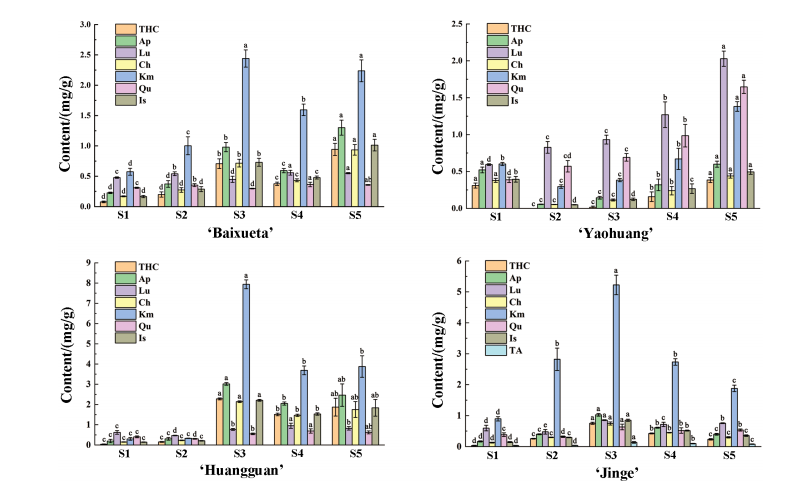
Figure 2. Flavonoid contents of tree peony cultivars at five blooming stages.
To explore the roles of PsMYB4 and PsEGL3 in yellow flower formation, qRT-PCR was performed across five floral stages in six tree peony cultivars. The results showed consistent expression patterns for both genes among all cultivars. Expression levels were higher in the darker yellow cultivars (‘High Noon’, ‘Huangguan’, and ‘Jinge’), and lower in the white, light yellow, and purple-red cultivars (‘Baixueta’, ‘Yaohuang’, and ‘Roufurong’). In ‘High Noon’ and ‘Huangguan’, both genes peaked at stage S4, showing a trend of increase followed by decline. In ‘Jinge’, PsMYB4 showed a fluctuating pattern, peaking at S5, while PsEGL3 increased steadily to S5. The distinct expression trends in ‘Jinge’ may be linked to its anthocyanin content. These findings suggest that PsMYB4 and PsEGL3 may contribute to anthocyanin suppression in yellow tree peony flowers.
Figure 3. Expression pattern analysis of PsMYB4 and PsEGL3 at five stages in the petals of six tree peony cultivars.
In preliminary research, the full-length sequences of PsMYB4 and PsEGL3 were cloned and analyzed. Sequence alignment showed that PsMYB4 contains a conserved R2R3 domain and a bHLH-interacting motif at the N-terminus, along with C-terminal C1 and C2 repression motifs. PsEGL3 possesses a bHLH domain and an N-terminal MYB-interacting region (MIR), which is essential for interaction with MYB transcription factors. Phylogenetic analysis revealed that PsMYB4 is an R2R3-MYB repressor closely related to Arabidopsis AtMYB4 (SG4 subgroup). PsEGL3 clustered within the GL3 clade of the bHLH subgroup IIIf and showed high similarity to several bHLH proteins involved in anthocyanin regulation, including AtGL3, AtEGL3, CpbHLH1, VvMYCA1, CsMYC2, and MdbHLH33.
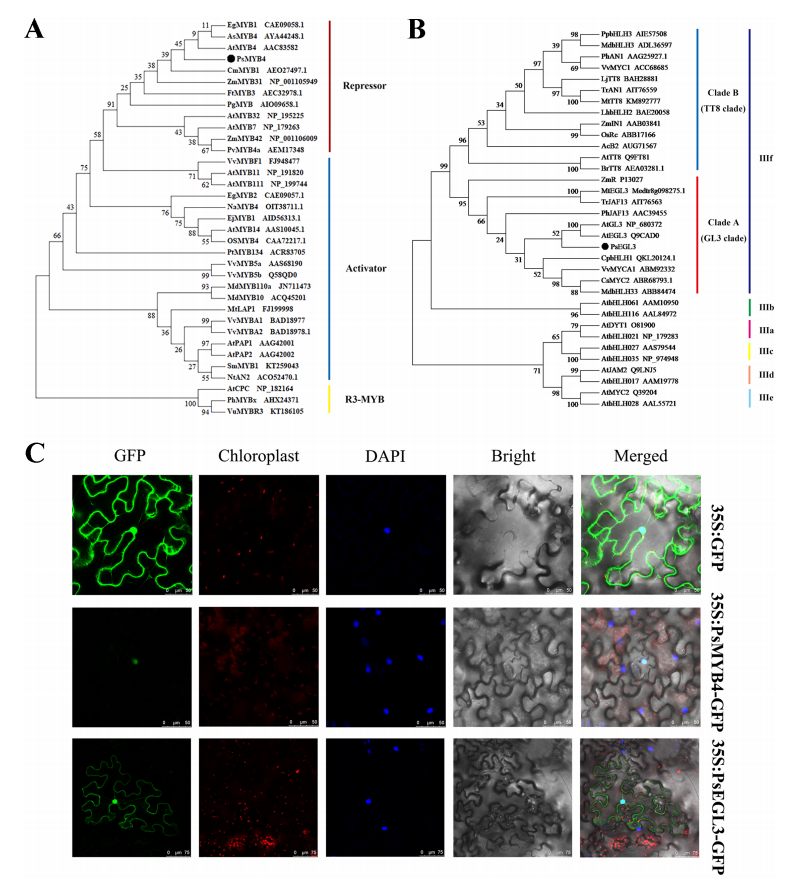
Figure 4. Phylogenetic tree analysis and subcellular localization of PsMYB4 and PsEGL3.
To investigate the roles of PsMYB4 and PsEGL3 in flavonoid biosynthesis, transgenic tobacco lines overexpressing each gene were developed. Two PsMYB4 lines (OE-PsMYB4-1 and OE-PsMYB4-2) and three PsEGL3 lines (OE-PsEGL3-1, OE-PsEGL3-2, and OE-PsEGL3-3) were obtained. Compared to the deep pink corolla of wild-type tobacco, OE-PsMYB4 lines showed a much lighter flower color, while PsEGL3 lines exhibited a slight lightening. Colorimetric analysis revealed significantly lower a and C values and higher L values in OE-PsMYB4 lines, indicating reduced red intensity and increased brightness. PsEGL3 lines showed variable color changes, with significant L value increases in two lines, and reduced a and C values only in OE-PsEGL3-3. Anthocyanin content was significantly reduced in PsMYB4 transgenic lines (p < 0.01) and moderately reduced in PsEGL3 lines (p < 0.05), consistent with the observed flower color changes. These results support the repressive role of both genes in anthocyanin biosynthesis.
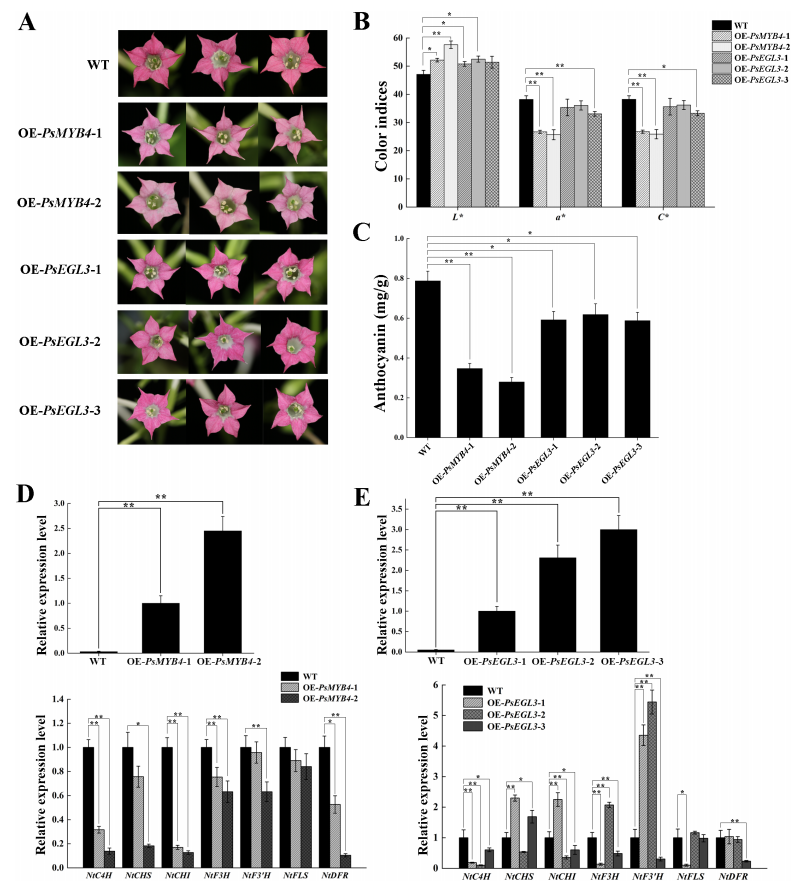
Figure 5. Functional verification of PsMYB4 and PsEGL3 in tobacco.
To further assess the roles of PsMYB4 and PsEGL3 in yellow flower formation, virus-induced gene silencing (VIGS) was used to suppress their expression in tree peony. TRV-PsMYB4 and TRV-PsEGL3 lines were generated, with TRV1 and TRV2 empty vector lines as controls. Yellow petals (with spots removed) were collected for analysis. In TRV-PsMYB4 and TRV-PsEGL3 lines, flower color became lighter, especially in TRV-PsMYB4. Flavonoid analysis showed significant decreases in compounds such as THC, apigenin, chrysoeriol, kaempferol, quercetin, and isorhamnetin in TRV-PsMYB4, and reduced levels of THC, apigenin, luteolin, chrysoeriol, and quercetin in TRV-PsEGL3, aligning with the observed petal color changes.
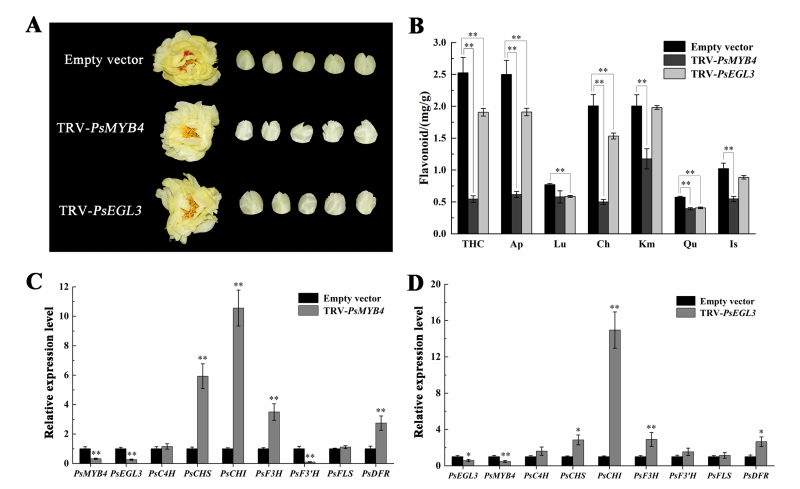
Figure 6. Functional verification of PsMYB4 and PsEGL3 in tree peony cultivar ‘High Noon’.
To verify the interaction between PsMYB4 and PsEGL3, yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays were conducted. In the Y2H assay, yeast cells co-expressing PsMYB4 and PsEGL3 grew normally on selective medium (SD-Leu-Trp-His-Ade), similar to the positive control, while the negative control showed no growth. These results confirm that PsMYB4 interacts with PsEGL3 in the yeast system.
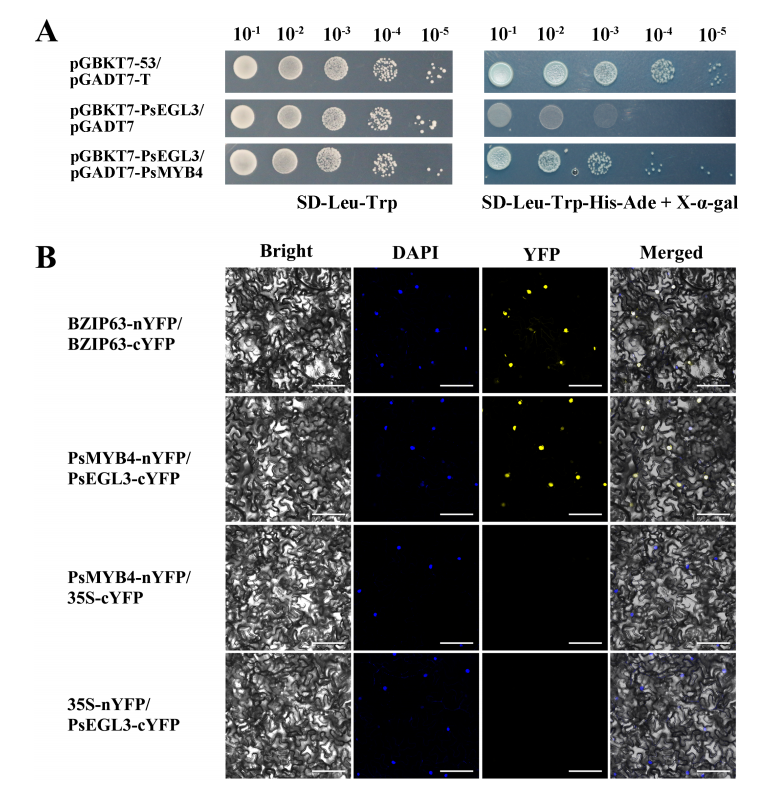
Figure 7. The protein interaction analysis between PsMYB4 and PsEGL3.
Overexpression and VIGS experiments suggest that PsMYB4 and PsEGL3 may work together to regulate flavonoid biosynthesis genes, particularly PsCHS, PsCHI, and PsDFR. To test this, promoters of these genes were isolated from ‘High Noon’ petals. Promoter analysis revealed multiple cis-elements recognized by MYB and bHLH transcription factors—such as MYB-binding sites, bHLH-binding sites, and G-box motifs—supporting the potential regulatory roles of PsMYB4 and PsEGL3 on these genes.
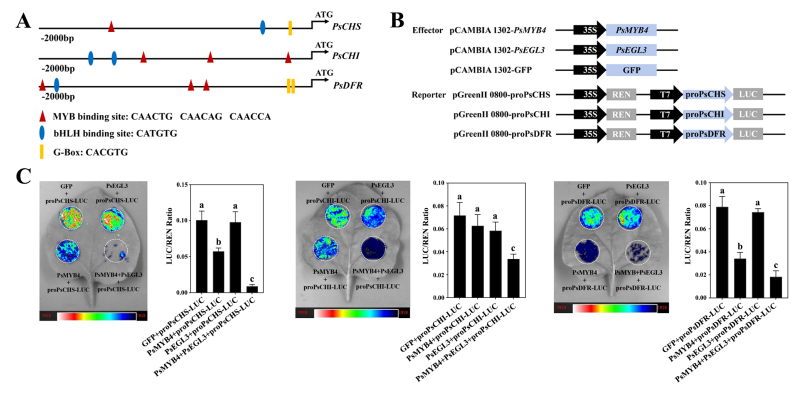
Figure 8. Transcription inhibition analysis of PsMYB4 and PsEGL3 against the promoters of PsCHS, PsCHI, and PsDFR of tree peony.
In summary, PsMYB4, an R2R3-MYB repressor, inhibits anthocyanin biosynthesis in tree peony yellow flowers. PsEGL3, a subgroup IIIf bHLH transcription factor, cooperates with PsMYB4 to suppress key flavonoid biosynthesis genes PsCHS, PsCHI, and PsDFR, thereby blocking the anthocyanin pathway. Alongside this, PsMYB111 promotes chalcone and flavonol production by activating PsCHS and PsFLS, shifting metabolic flux toward flavonol synthesis. Together, these regulatory mechanisms drive the formation of yellow pigmentation in tree peony petals. These insights deepen understanding of yellow flower development and may aid in breeding new yellow cultivars with enhanced ornamental value.
Related News
2025-07-10
2025-07-08
2025-07-03
Literature Sharing | Regulation of co-translational mRNA decay by PAP and DXO1 in Arabidopsis
2025-07-01
2025-06-27
2025-06-24
2025-06-20



