Literature Sharing | Putative Upstream Regulators DoNF-YB3 and DoIDD12 Correlate with DoGSTF11 Expression and Anthocyanin Accumulation in Dendrobium officinale
Release time:
2025-07-10
This study explores the role of the DoGSTF11 gene in Dendrobium officinale, a traditional medicinal herb. While previous research has focused on polysaccharides and alkaloids, this work addresses the lesser-known anthocyanin pathway. The researchers found that DoGSTF11 is highly expressed in the purplish variety of D. officinale and is localized in the nucleus and cell membrane, though it lacks transcriptional activation ability. Overexpressing DoGSTF11 in tomato led to increased anthocyanin accumulation, suggesting it plays a role in anthocyanin transport or sequestration. Protein interaction studies revealed that DoGSTF11 interacts with DoGST31, and that DoIDD12 and DoNF-YB3 may regulate its expression. Overall, the findings highlight DoGSTF11 as a positive regulator of anthocyanin accumulation and offer new insights for flavonoid metabolic engineering in D. officinale.

Two color variants of Dendrobium officinale—green and purplish—were used for analysis. Microscopy showed that anthocyanins in the purplish stems mainly accumulated in the epidermal cells, while the green stems showed little to no pigment. Quantitative analysis confirmed that the purplish variety had the highest anthocyanin content.
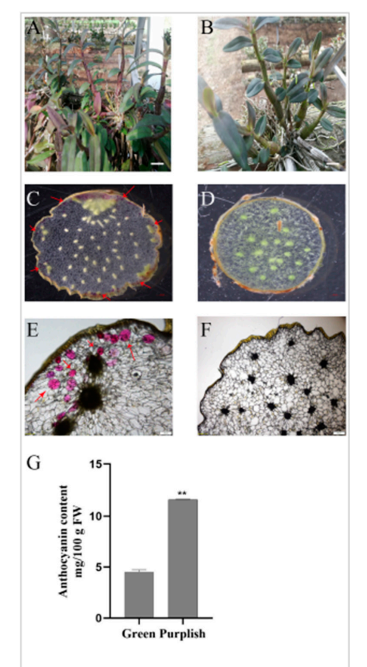
Figure 1. Phenotypic observation of different Dendrobium officinale varieties
To investigate the role of GST genes in anthocyanin regulation in D. officinale, expression analysis was conducted across green and purplish phenotypes. Eleven DoGST genes showed significant differential expression, with DoGSTF11 displaying the most prominent change. Phylogenetic analysis revealed that DoGSTF11 clusters with known anthocyanin-related GSTs, such as Arabidopsis TT19, while other DoGSTs formed a separate group. Sequence alignment showed that DoGSTF11 shares high similarity with these anthocyanin-related GSTs, especially grape VvGST4 (67% identity). These results suggest DoGSTF11 may play a key role in anthocyanin synthesis and metabolism.
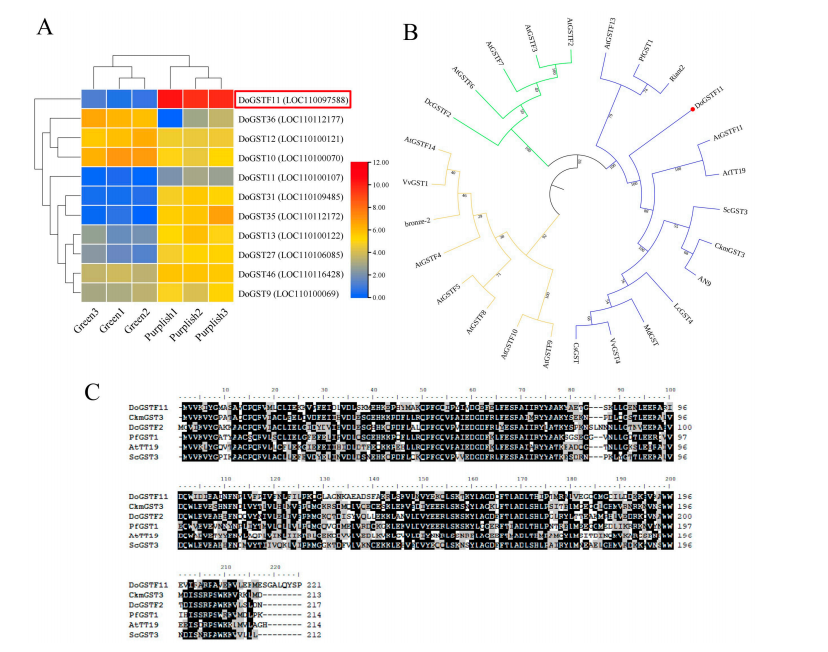
Figure 2. DoGSTs expression and phylogenetic analysis.
Subcellular localization of DoGSTF11 was examined by transiently expressing a DoGSTF11-GFP fusion in tobacco leaves. Confocal microscopy showed that the GFP signal from both the control (35S:GFP) and DoGSTF11-GFP was present in the cell membrane and nucleus, indicating that DoGSTF11 localizes to these two cellular compartments.
To assess the function of DoGSTF11, it was stably overexpressed in tomato seedlings. Three transgenic lines were generated, and their stems and leaves showed noticeable purple pigmentation, especially in line OE3, which had the highest DoGSTF11 expression. Anthocyanin measurements confirmed significantly increased levels in all transgenic lines compared to wild type, with OE3 displaying the highest accumulation. These findings suggest that DoGSTF11 positively regulates anthocyanin synthesis.
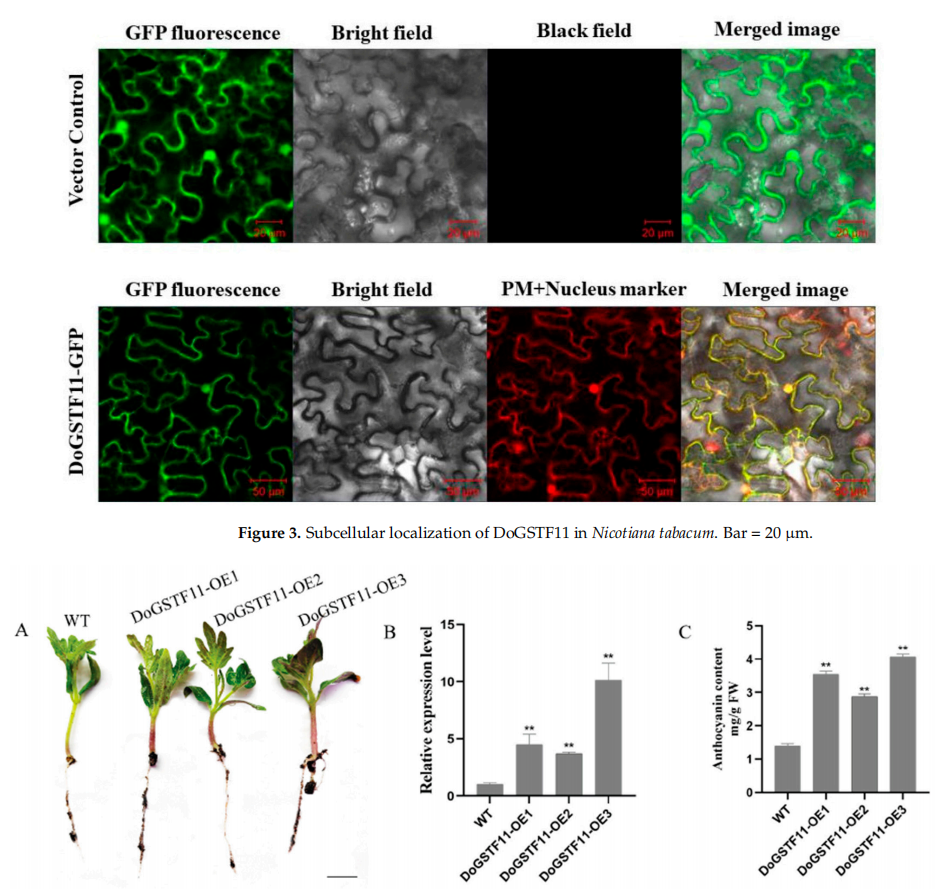
Figure 4. Heterologous expression of DoGSTF11 in transgenic Solanum lycopersicum.
The transcriptional self-activation activity of DoGSTF11 was analyzed. As shown in Figure 5A, the negative control (pGBKT7) was able to grow on SD/-Trp plates but failed to grow on SD/-Trp/-His, SD/-Trp/-His/-Ade, and SD/-Trp/-His/-Ade + X-α-gal plates. The experimental group showed the same growth pattern as the negative control, indicating that DoGSTF11 lacks transcriptional self-activation activity.
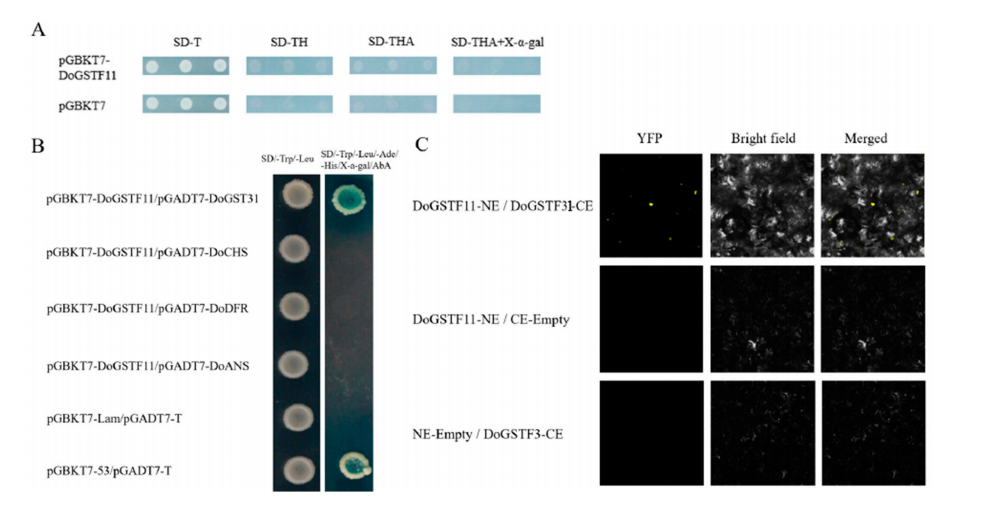
Figure 5. Analysis of DoGSTF11-interacting proteins.
To identify upstream regulators of DoGSTF11, a pHIS2-DoGSTF11pro bait plasmid was constructed for yeast one-hybrid screening. Sequence analysis identified 21 unique positive clones after removing empty vectors and duplicates. These clones were able to grow on selective media containing 3-AT, indicating promoter-binding activity. Further analysis suggested that DoIDD12 and DoNF-YB3 are potential upstream transcriptional regulators of DoGSTF11.
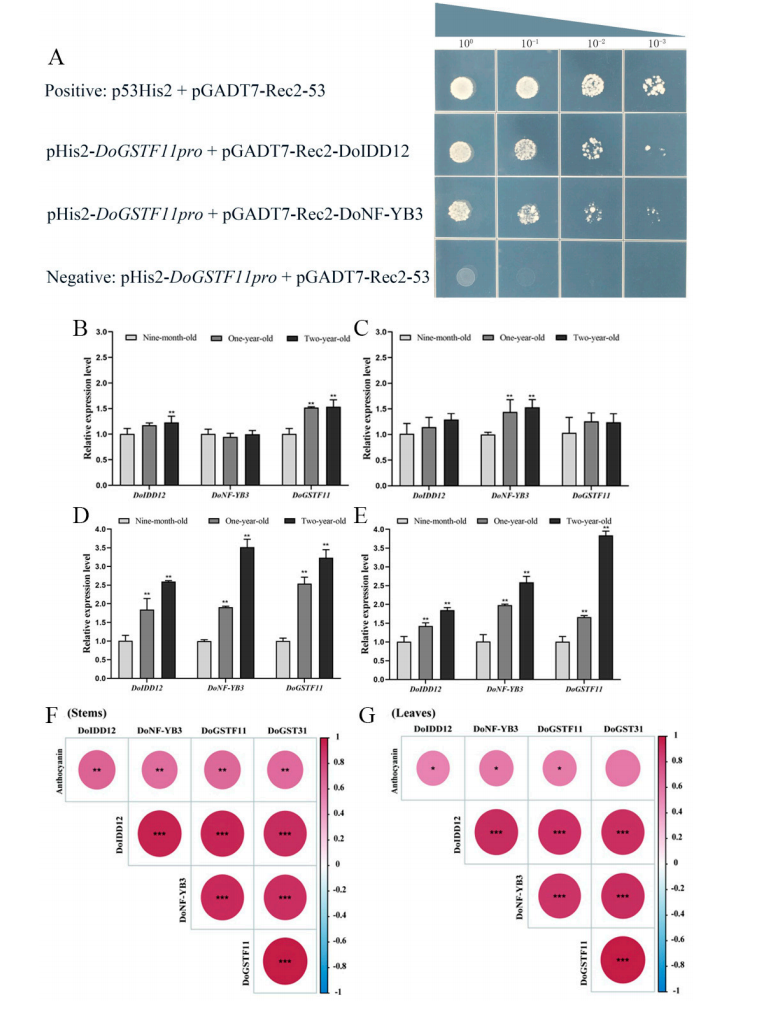
Figure 6. Screening of upstream regulatory genes of DoGSTF11.
This study identifies DoGSTF11 as a key positive regulator of anthocyanin accumulation in purple-stem Dendrobium officinale. Protein interaction assays (Y2H and BiFC) showed that DoGSTF11 interacts with DoGST31, promoting anthocyanin buildup. Upstream regulators DoIDD12 and DoNF-YB3 were also identified by Y1H assays. A proposed model suggests that DoGSTF11 enhances anthocyanin synthesis in the endoplasmic reticulum and facilitates its transport to the vacuole for accumulation. These findings deepen understanding of the molecular mechanisms driving anthocyanin biosynthesis in this medicinal plant.
Related News
2025-07-10
2025-07-08
2025-07-03
Literature Sharing | Regulation of co-translational mRNA decay by PAP and DXO1 in Arabidopsis
2025-07-01
2025-06-27
2025-06-24
2025-06-20



