Dual Luciferase Reporter Assay Services
Dual Luciferase Reporter Assay Service for Gene Expression and Regulatory Element Analysis
Service Overview
Genetic reporter gene systems are now widely used in eukaryotic gene expression and cell physiology studies. However, single-reporter assays are often affected by various experimental conditions. To improve accuracy, dual-reporter gene systems are commonly employed.
A dual-reporter system refers to the simultaneous expression and measurement of two independent luciferase enzymes within the same experimental setup. Typically, the experimental reporter reflects the specific biological response being studied, while a co-transfected control reporter provides a baseline. This strategy effectively minimizes the influence of external variables and enhances the overall data reliability.
Technical Principle
This system utilizes Firefly luciferase and Renilla luciferase, both of which catalyze oxidation reactions that emit bioluminescence. The intensity of the fluorescence directly reflects the expression levels of the two enzymes. Firefly luciferase catalyzes the oxidation of luciferin to produce oxyluciferin, emitting yellow-green light in the process. Renilla luciferase oxidizes coelenterazine to generate coelenteramide, which emits blue light.
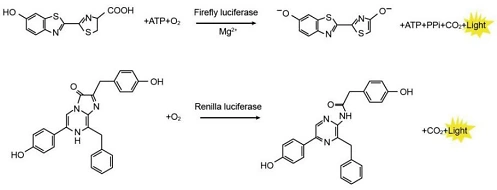
To assess gene regulation, the regulatory element of the target gene is cloned upstream or downstream of the Firefly luciferase gene to construct a luciferase reporter plasmid. This plasmid is then transfected into cells. After appropriate treatment or stimulation, the cells are lysed, and luciferase activity is measured. Changes in Firefly luciferase activity indicate the regulatory effects of the treatment or condition on the target gene.
At the same time, Renilla luciferase is used as an internal control. Both reporter genes are constructed within the same plasmid and driven by separate promoters. This configuration helps eliminate variations caused by transfection efficiency or differences in basal transcriptional activity between samples.
Advantages of the Dual Reporter Assay
In Vivo Assays: Endogenous expression allows a more accurate representation of in vivo molecular interactions.
High Sensitivity: Not affected by interference from other intracellular components.
High Accuracy: No endogenous expression of luciferase reporters in either plant or mammalian systems, ensuring clean background and reliable results.
Applications Scenarios
▷ Validation of miRNA–Target Gene Interaction
miRNAs regulate target genes through cleavage or translational inhibition, precisely controlling their expression levels.
By inserting the 3' UTR of the target gene into the Firefly luciferase (LUC) reporter vector, the binding of miRNA to the inserted sequence suppresses luciferase translation, resulting in decreased fluorescence intensity.
▷ Targeted Interaction between miRNA and lncRNA/circRNA
Many lncRNAs structurally resemble mRNAs, allowing miRNAs to negatively regulate lncRNAs via mechanisms similar to mRNA regulation.
By inserting candidate lncRNA sequences into the luciferase reporter vector, miRNA binding inhibits luciferase translation and reduces fluorescence, enabling detection of miRNA-lncRNA or miRNA-circRNA interactions.
▷ Transcription Factor Regulation of Downstream Genes
Transcription factors bind to specific cis-regulatory elements within promoter regions to regulate gene expression.
By inserting promoter sequences into the LUC reporter vector and co-transfecting the transcription factor, fluorescence increases if binding occurs, indicating transcriptional activation.
▷ Promoter Structure Analysis
Promoter regions can be truncated or mutated at specific sites and then cloned into luciferase reporter vectors to evaluate their transcriptional activity.
▷ Promoter SNP Analysis
Single nucleotide polymorphisms (SNPs) within promoter regions can influence gene expression.
The luciferase assay enables quantitative comparison of promoter activity associated with different SNP alleles.
▷ Verification of Promoter Activity
To validate promoter function, the test promoter is cloned upstream of the luciferase gene, and reporter activity is measured to assess promoter strength.
▷ Verification of Transcription Factor Activity
The transcription factor of interest is fused with a GAL4 DNA-binding domain in a single plasmid.
This construct is co-transfected with a Firefly luciferase reporter driven by a GAL4-TATA regulatory element.
Changes in fluorescence intensity reflect whether the transcription factor exhibits activation or repression function.
Sample Requirements
| Application Scenario | Requirements |
| Interaction between miRNA and Target Gene | miRNA sequence information, species information; Target gene sequence information, species information, gene name |
| Targeted Interaction between miRNA and lncRNA/circRNA | miRNA, lncRNA/circRNA sequence information, species information |
| Effect of Transcription Factors on Downstream Genes | DNA sequence information, species information; Target gene sequence information, species information, gene name |
Service Details
Plant System:
Service Process | Working Days | Deliverables |
Order placement | 10 | 1. One aliquot of the recombinant plasmid |
Experimental Material Confirmation | ||
Gene Synthesis / Vector Construction | ||
Tobacco Injection | 10 | |
Luciferase Assay | ||
Quantitative Analysis | 5 | |
Data Processing | ||
Total | 25 |
293T System:
Service Process | Working Days | Deliverables |
Order placement | 10 | 1. Plasmid from gene synthesis/vector construction and sequencing validation report |
Experimental Material Confirmation | ||
Gene Synthesis / Vector Construction | ||
293T cell transfection | 10 | |
Luciferase Assay | ||
Quantitative Analysis | 5 | |
Data Processing | ||
Total | 25 |
Case Results Study
Tobacco Transient Expression System
To evaluate the DNA-binding capability of Protein A in vivo, we conducted a dual luciferase assay using a tobacco transient expression system.
The results showed that the relative LUC/REN activity of the pro-LUC + Protein A group was significantly higher than that of the pro-LUC + empty vector group, indicating that Protein A binds to the promoter.
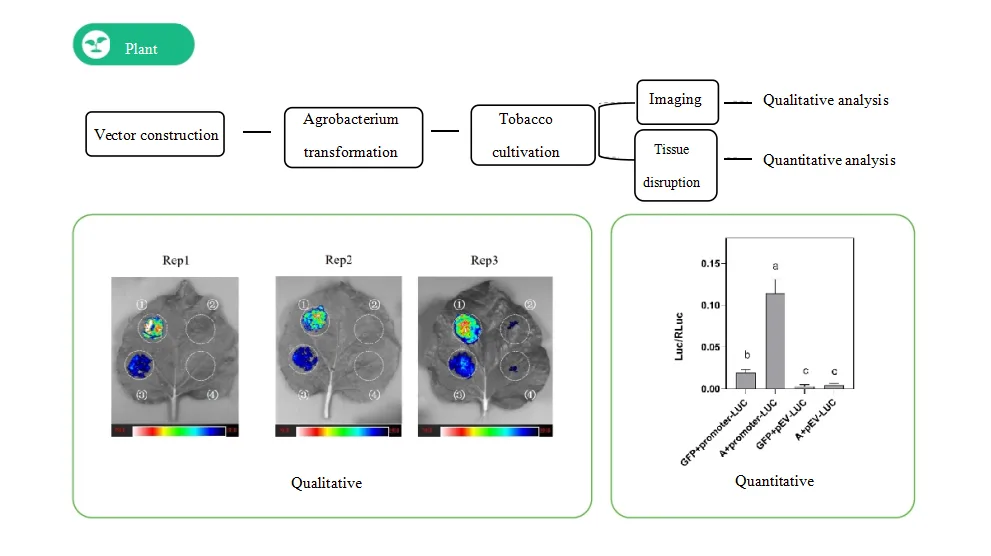
Interaction Between Protein A and the Promoter in Tobacco
In this dual-luciferase reporter assay, tobacco leaves were co-infiltrated with pro-LUC + Protein A and pro-LUC + empty vector.
Luciferase activity (LUC/REN) was calculated, and statistical significance was determined using Student's t-test (P < 0.01).
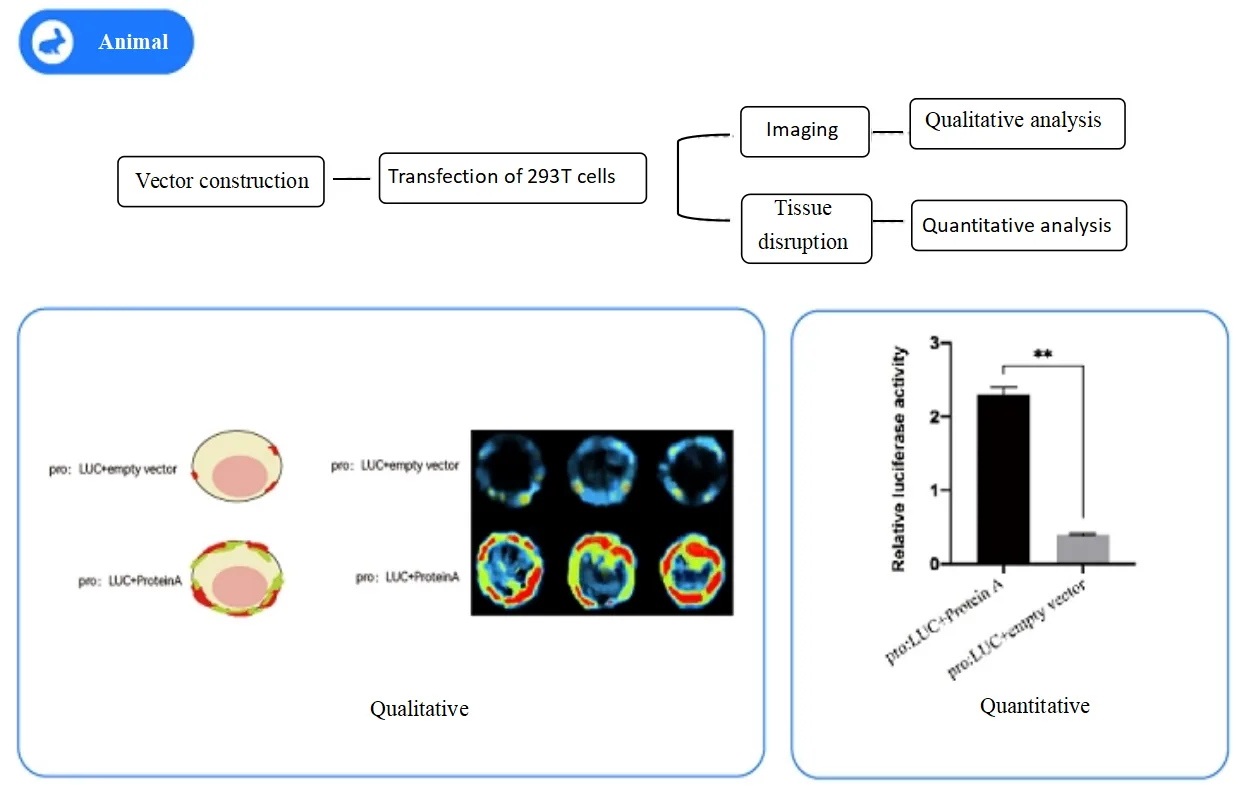
HEK-293T Cell System
To further validate the DNA-binding activity of Protein A, we performed a dual luciferase assay in HEK-293T cells.
The relative LUC/REN activity of the pro-LUC + Protein A group was significantly higher than that of the pro-LUC + empty vector group, confirming that Protein A binds to the promoter in mammalian cells as well.
Frequently Asked Questions (FAQs)
1. What if the luciferase signal is too high or too low?
An ideal fluorescence signal should fall within the 5- to 6-digit range. If the signal is too high or too low, try the following:
(1) Signal Too High:
a. Reduce the amount of transfected plasmid.
b. After cell lysis, centrifuge and measure the supernatant, or dilute the lysate before testing.
(2) Signal Too Low:
a. Low transfection efficiency: Optimize transfection conditions and ensure high-quality plasmid DNA.
b. Poor lysis efficiency: Avoid prolonged cell culture as this can reduce lysis efficiency. Ensure sufficient lysis buffer is used, and maintain a lysis duration of 24–48 hours.
c. Suboptimal assay conditions: All components, including lysate and substrate working solution, should be at room temperature during the assay.
d. Substrate oxidation failure: Protect substrates from light and seal containers tightly.
· Firefly luciferase substrate should be stored at –20°C.
· Renilla luciferase substrate is recommended to be stored at –80°C.
· Prepare working solutions fresh before use.
2. What if replicate variability is high?
Luciferase reporter assays are highly sensitive and can be affected by many factors.
In addition to using Renilla luciferase as an internal control, it is recommended to set up at least three biological replicates to minimize variability. Differences within the same order of magnitude are generally acceptable.
a. Centrifuge lysates after lysis to ensure sample uniformity.
b. Ensure precise pipetting by regularly calibrating pipettes.
3. What is the difference between Firefly and Renilla luciferase?
Luciferase Type | Cofactors Required | Emission Range | Recommended Detection Wavelength |
|---|---|---|---|
Firefly luciferase | O₂, ATP, Mg²⁺ | 540–600 nm (green-yellow) | 560 nm |
Renilla luciferase | O₂ only | 460–540 nm (blue) | 460 nm |
Ready to Explore the Power of Dual Luciferase Reporter Assays?
Unlock the potential of your gene expression analysis with our Dual Luciferase Reporter Assay System. Whether you're studying miRNA interactions, transcription factor activity, or SNP analysis, our reliable and sensitive system can help advance your research. Contact us today to learn more or to place an order!
Disclaimer: All products and services are for research use only and are not intended for diagnostic or therapeutic applications.
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry