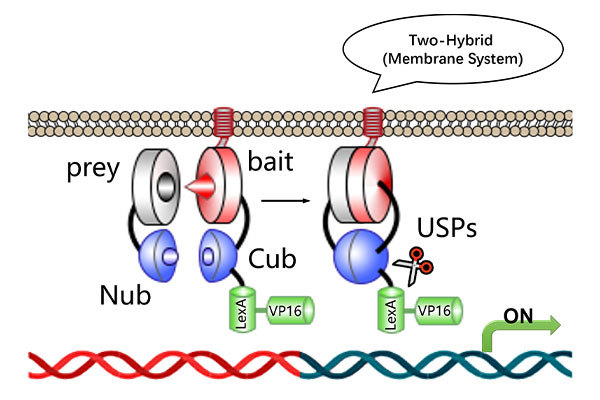
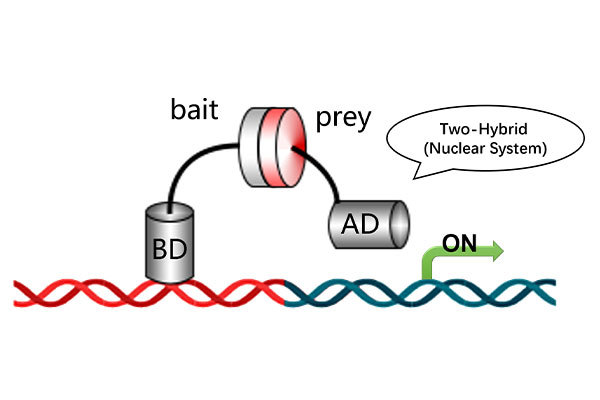
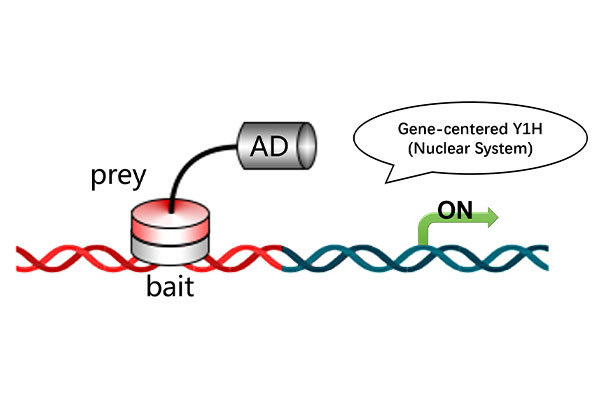
Services
Providing Advanced Biological Technical Services for Researchers in Fundamental Science
TF-Centered Yeast One-Hybrid (Y1H-TF) Screening Service
Service Overview
The TF-centered Yeast One-Hybrid (Y1H-TF) Screening Service is a cutting-edge solution for identifying transcription factor (TF) recognition elements. Using a comprehensive yeast motif library of 50,000 sequences, this service enables the efficient discovery of 7-bp cis-acting elements that directly interact with your TF of interest. This approach streamlines motif exploration and validation processes, delivering high-confidence results around just one month.
Applications:
· Identification of Core Regulatory Elements: Uncover key TF-DNA interaction sites to understand transcription regulation.
· Efficient Discovery of Novel Cis-Acting Elements: Reduce false positives while identifying critical motifs for TF interactions.
· Supports Gene Regulatory Network Studies: Combine with ChIP or transcriptome data to map out downstream genes and pathways.
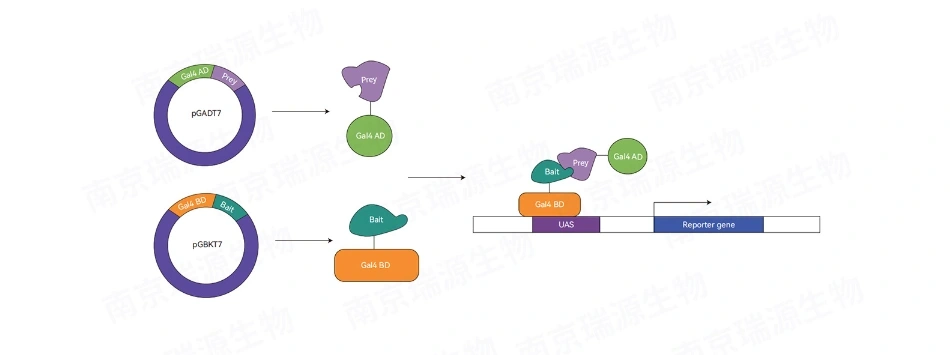
Why Choose Our TF-Centered Y1H Service?
· Comprehensive Coverage: Our random motif library includes all possible 7-bp sequences derived from ATCG combinations, ensuring compatibility with any species.
· Rapid & Reliable Results: Identify 3-5 high-confidence motifs after filtering redundancies for a streamlined discovery process.
· One-Step Workflow: No need for complex genetic transformations or antibody preparations. We provide direct results with minimal steps.
· Cost & Time Efficiency: Complete your project around one month, saving significant time and resources compared to traditional ChIP or transgenic methods.
· Advanced Bioinformatics Integration: Enhance your analysis with genome scanning, ChIP data, or transcriptome analysis to locate downstream regulatory genes.
Service Workflow and Deliverables
| Service Content | Working Days | Deliverables |
|---|---|---|
| 1. Gene Synthesis (Codon Optimization) / Vector Construction | 10 | 1. Positive clone sequencing results (depending on sequencing results) 2. Word document project report (including experimental data) 3. Genome scan analysis 4. Bait plasmid (Optional) 5. Y1H verification motif plasmid (Optional) |
| 2. Bait Plasmid Self-activation Testing | 7 | |
| 3. Co-transformation of Bait Plasmid and Library Plasmid | 10 | |
| 4. Yeast Screening Condition Optimization | ||
| 5. Sequencing of Screening Results | ||
| 6. NEW PLACE Analysis and Organization | 5 | |
| 7. One-to-one Validation of Identified Motifs | ||
| 8. Revalidation of Unidentified Motifs | ||
| 9. Interaction Motif Promoter Scan Analysis | 5 | |
| 10. Data Organization and Analysis | ||
| 11. Determining Motif Boundaries (Optional) | 17 |
Technical Details
Screening Methodology
Library Construction: Randomized 7-bp sequences cloned into yeast motif libraries.
Screening: Yeast transformation with TF-coding bait plasmid followed by stringent selection using 3-AT competitive inhibitors.
Validation: Positive clones undergo sequencing and reverse validation to ensure reliability.
Key Protocols
Background Screening: 75 mM 3-AT concentration ensures minimal noise in results.
Yeast Transformation: Uses proprietary techniques for high-efficiency plasmid integration and selection.
Integrative Applications
With ChIP Data: Enhances the localization of TF-binding motifs on promoters.
With Transcriptome Data: Links identified motifs to genes regulating specific phenotypes or pathways.
Why Researchers Prefer TF-Centered Y1H?
Traditional methods like ChIP or EMSA often require significant time, resources, and expense. Our TF-centered Y1H service offers:
A Complete Solution around One Month: Discover high-confidence motifs efficiently.
Cost-Effective: Ideal for researchers without transgenic systems or plant-specific ChIP-grade antibodies.
An Integrative Platform: Combines wet lab and bioinformatics analyses for robust and accurate results.
FAQs of TF-Centered Y1H
· What species are covered in the motif library?
Our motif library includes all possible 7-bp sequences derived from ATCG combinations, ensuring it covers motifs for any species.
· How many sequences can be identified in one screening?
Typically, dozens of sequences are found, but after removing redundancies, 3-5 unique high-confidence motifs are delivered.
· How do I use the 7-bp motif to identify downstream regulatory genes?
With ChIP Data: Combine our results with ChIP data to locate promoter regions interacting with the identified motifs.
With Transcriptome Data: Scan promoters of differentially expressed genes for occurrences of our identified motifs, linking TFs to specific regulatory genes.
· Why are two primers containing insertion sequences synthesized?
A total of 3 × 47, or 49,512 possible sequences, are covered by these primers to ensure comprehensive motif discovery.
· Why is the random sequence library constructed with only 7-bp sequences?
Determining the boundaries of new cis-elements requires additional effort. Longer insertion sequences would increase the workload for identifying these boundaries. Using 7-bp sequences helps reduce the complexity and accelerates the process.
· Why are random sequences inserted into the SmaI restriction site?
The SmaI enzyme recognizes the sequence "CCCGGG", which conveniently serves as a marker for identifying the inserted sequence. This results in a structure of "CCCHNNNNNNCGGG", where CCC and GGG act as the marker sites.
Case Study
Screening of Random DNA Library Using AtbZIP53 Protein
(Plant Mol Biol. 2014, 86:367-380)
bZIP transcription factors are a crucial class of plant transcription factors that play a significant role in plant growth and development. While only a few bZIP family members have been studied, it has been found that they mainly bind to core cis-acting elements, such as “ACGT”. However, it remains unclear whether these factors recognize additional cis-elements and thus exert their biological functions.
In this study, we used AtbZIP53 to screen a random DNA library and successfully identified one unknown element and five known DNA sequences.
Identification of the New Cis-Element Recognized by AtbZIP53
We determined that AtbZIP53 specifically recognized the new cis-element "GTGCG" in the DNA sequences.
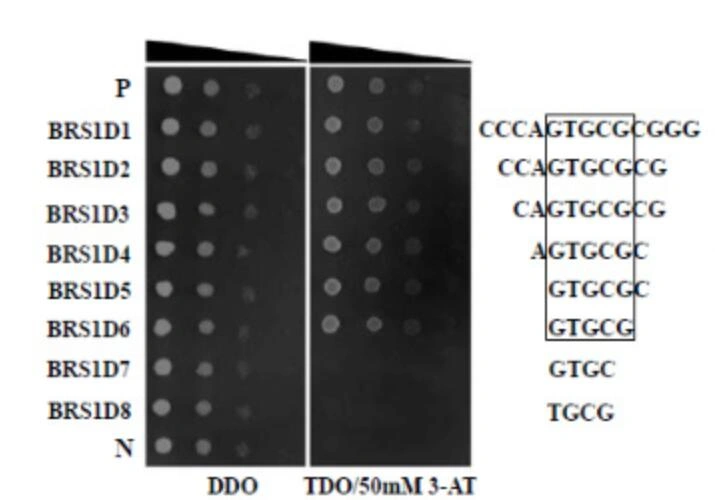
Yeast One-Hybrid Validation
The interaction between AtbZIP53 protein and different cis-elements was validated using Yeast One-Hybrid (Y1H) technology, ensuring the specificity of the recognition.

Ready to Accelerate Your Transcription Factor Research?
Unlock new possibilities with our TF-Centered Yeast One-Hybrid Screening Service! Contact us today to discuss your project requirements and receive a customized solution. When inquiring about this service, please reference Service Code: SRY8012.
Statement: Services sold by our company are intended for research purposes only and must not be used for diagnostic or therapeutic purposes.
Related Product: Yeast One Hybrid (TF-Centered) Vector Kit [CAT:RY8012]
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry

Contact Us

Tel: +86 025-85205672

Email: info@pronetbio.com

Address: Building 3C, Nanjing Xianlin Zhigu,
Qixia District, Nanjing, Jiangsu, China, 210033

Need more info?
Let's connect!