Services
Providing Advanced Biological Technical Services for Researchers in Fundamental Science
CRISPR-Cas12a-Based Yeast Genome Editing Service
Yeast Genome Editing with CRISPR-Cas12a – Knockout, Knock-in, and Multiplex Editing
Overview
ProNet Biotech introduces a highly flexible and efficient CRISPR-Cas12a-based yeast genome editing platform, enabling single-gene knockout, multi-gene editing, and targeted gene insertion in Saccharomyces cerevisiae and other yeast strains. Compared to traditional TALEN and ZNF technologies, the CRISPR-Cas12a system offers faster workflows, higher precision, and lower off-target effects, making it ideal for advanced functional genomics, synthetic biology, and strain engineering.
Technical Background
CRISPR/Cas12a enables genome editing through a three-step process: target recognition, cleavage, and repair. The system transcribes a pre-crRNA that is processed by Cas12a into a mature crRNA, forming a nuclease complex. This complex locates a 5'-TTN-3' PAM site, binds the target via base pairing, and performs a staggered double-strand break using its RuvC domain, generating sticky ends (4–5 nt).
Repair is triggered via two major mechanisms:
NHEJ (Non-Homologous End Joining), leading to frameshift mutations
HDR (Homology-Directed Repair), used for precise gene insertions when donor DNA with ~500–1000 bp homology arms is provided
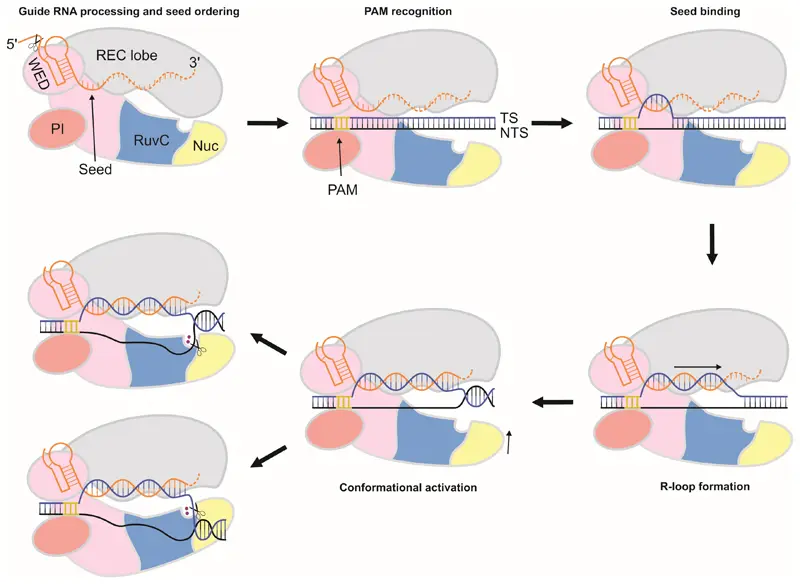
Fig1. CRISPR/Cas12a Gene Editing Principles (Swarts DC al et, 2017)
Application Areas
Functional gene disruption and loss-of-function studies
Rapid generation of genome-edited strains for phenotype analysis or drug screening
Multi-gene pathway analysis and regulatory mechanism exploration
Metabolic engineering and industrial yeast optimization
Yeast Genome Editing Workflow
|
Project Type |
Working Days |
Key Steps |
|---|---|---|
|
Knockout / Knock-in Editing |
45 days |
1. Target site design |
Deliverables
Glycerol stocks and/or plates of edited strains
Sequencing files confirming edit accuracy
One electronic experiment report including raw data
Key Advantages
Precision Editing: High fidelity with Cas12a system, reduced off-target risks
Edit Validation: Includes colony PCR and sequencing for confirmation
Flexible Design: Accepts user-defined target sequences or provides sgRNA design
Fast Turnaround: 30–50% faster than conventional gene editing workflows
Cas12 Vector Library: Pre-validated Cas12a vectors for common yeast strains
Multi-Application Ready: From academic research to industrial strain improvement
Sample Requirements
Digital Data (Required for all projects):
· Target gene name and ID
· Full genome sequence of the yeast strain to be edited
Physical Materials (if applicable):
|
Sample Type |
Submission Standard |
Shipping Condition |
|---|---|---|
|
Yeast strain |
≥1×10⁸ CFU/mL, contamination-free |
Room temp, deliver within 48 hrs |
|
Plasmid (sgRNA vector) |
≥100 ng/μL, ≥20 μL |
Ice pack, within 72 hrs |
|
Glycerol stock |
≥500 μL, labeled with antibiotic resistance info |
Dry ice, 1–3 days |
|
Plate colonies |
Contamination-free |
Room temperature |
Validation Criteria
1. PCR confirmation of edit at target site (gel image included)
2. Sanger sequencing alignment confirming edit accuracy
3. Delivery of edited strain and experimental report with original data
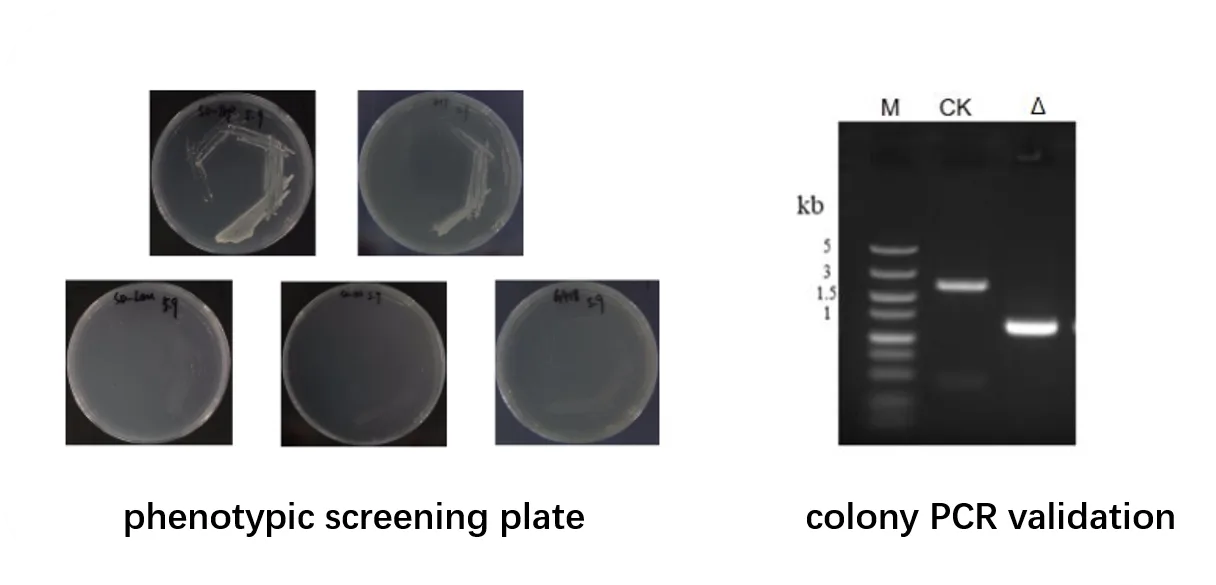
Representative Data
Phenotype screening images of edited strains
Electroporation plate growth results
PCR validation gel images
Sanger sequencing trace alignment files
Ordering Information
For pricing, quote requests, or custom project consultation, please email us at info@pronetbio.com or leave a message below.
References
-
Li ZH, Liu M, Lyu XM, Wang FQ, Wei DZ. CRISPR/Cpf1 facilitated large fragment deletion in Saccharomyces cerevisiae. J Basic Microbiol. 2018;58(12):1100-1104.
-
Li, Z. H., Liu, M., Wang, F. Q., & Wei, D. Z. (2018). Cpf1-assisted efficient genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. Biotechnology letters, 40(8), 1253–1261.
-
Verwaal, R., Buiting-Wiessenhaan, N., Dalhuijsen, S., & Roubos, J. A. (2018). CRISPR/Cpf1 enables fast and simple genome editing of Saccharomyces cerevisiae. Yeast (Chichester, England), 35(2), 201–211.
-
Swiat, M. A., Dashko, S., den Ridder, M., Wijsman, M., van der Oost, J., Daran, J. M., & Daran-Lapujade, P. (2017). FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic acids research, 45(21), 12585–12598.
-
Chen, B. C., Chen, Y. Z., & Lin, H. Y. (2023). An Introduced RNA-Only Approach for Plasmid Curing via the CRISPR-Cpf1 System in Saccharomyces cerevisiae. Biomolecules, 13(10), 1561.
Related Service: Yeast Gene Knockout Service Based on Homologous Recombination.
Important Notes: This service is intended for research use only. It is not for diagnostic or therapeutic purposes.
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry

Contact Us

Tel: +86 025-85205672

Email: info@pronetbio.com

Address: Building 3C, Nanjing Xianlin Zhigu,
Qixia District, Nanjing, Jiangsu, China, 210033

Need more info?
Let's connect!