BSA Analysis and Candidate Gene Validation Service
BSA Service: Comprehensive Trait Mapping and Candidate Gene Verification
Overview
ProNet Biotech offers a comprehensive BSA (Bulked Segregant Analysis) and candidate gene validation service utilizing advanced Illumina sequencing platforms. Our end-to-end solution covers library construction, sequencing, BSA analysis, gene mining, and functional validation. Researchers only need to provide the original materials, and we deliver fast, accurate identification of trait-associated genomic regions, followed by candidate gene validation through techniques such as qRT-PCR, EMSA, Yeast One-Hybrid (Y1H), and Yeast Two-Hybrid (Y2H) assays.
Technical Principle
Bulked Segregant Analysis (BSA) is a powerful approach to identify genomic regions linked to specific traits. Two parent lines exhibiting extreme phenotypes are crossed to generate F1 progeny, which are then selfed to produce an F2 segregating population. Individuals with extreme phenotypes are selected from the F2 population and grouped into two DNA bulks. High-throughput sequencing is used to compare allele frequency differences (AF) between bulks, enabling the identification of candidate regions associated with the target trait.
Applications
· Gene Mapping: Identify genomic loci associated with specific phenotypes.
· Candidate Gene Mining: Discover genes potentially responsible for trait variation.
· Marker Development: Develop molecular markers linked to important traits for breeding.
· Functional Studies: Design downstream experiments to validate candidate gene functions.
Research Focus: Yield traits, disease resistance, stress tolerance, and other agronomically important characteristics in plants and animals.
Key Advantages
High Efficiency: Rapid and precise detection using both ΔSNP Index and ED methods.
Comprehensive Verification: In-house qRT-PCR, EMSA, Y1H, and Y2H validation services to confirm candidate gene functions.
End-to-End Service: From material collection to sequencing, analysis, and functional validation.
Rigorous Quality Control: Base quality distribution, sequencing accuracy checks, and extensive data evaluation.
Tailored Support: Tailored workflows based on genome size, sample type, and research objectives.
Service Workflow
Our BSA analysis and candidate gene validation service follows a comprehensive and streamlined workflow to ensure high-quality and timely results.
Workflow Overview
Research Material Preparation → DNA Extraction → Library Construction and Sequencing → Raw Data Acquisition → Sequencing Data Quality Control → Alignment to Reference Genome → SNP and Indel Variant Detection → Trait-Associated Region Mapping → Candidate Gene Identification and Functional Annotation → Candidate Gene Functional Validation (Optional)
Service Timeline
|
Step |
Description |
Estimated Working Days |
|---|---|---|
|
1 |
Library Construction and Sequencing |
20 |
|
2 |
Sequencing Data Quality Control |
1 |
|
3 |
Alignment to Reference Genome |
2 |
|
4 |
Variant Detection, Annotation, and Plotting |
7 |
|
5 |
Candidate Gene Mining, Annotation, and Plotting |
4 |
|
6 |
Data Organization and Delivery |
1 |
|
7 |
Candidate Gene Functional Validation |
Based on requirements |
Note: The timeline for candidate gene validation depends on the specific validation methods selected (e.g., qRT-PCR, EMSA, Y1H, Y2H) and project complexity.
Candidate Gene Validation
Following BSA analysis, we provide robust experimental validation for candidate genes using:
· qRT-PCR (quantitative expression validation)
· EMSA (protein-DNA interaction verification)
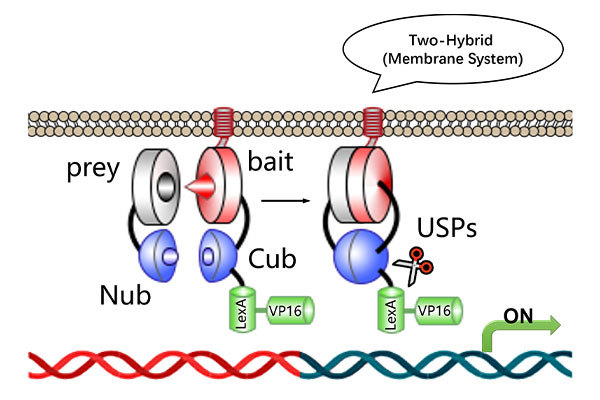
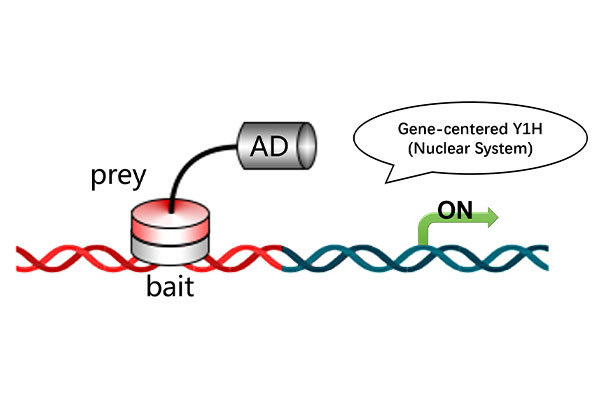
· Yeast One-Hybrid (Y1H) (transcriptional activation assays)
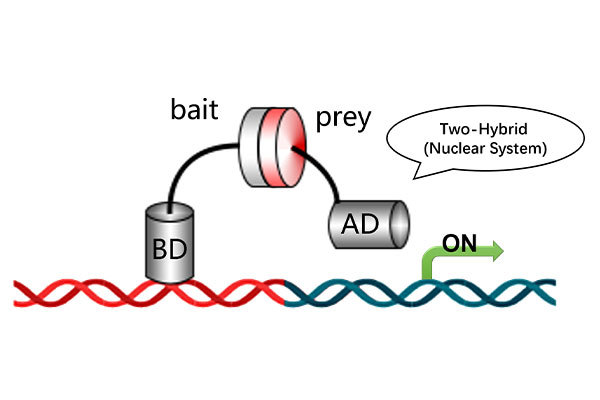
· Yeast Two-Hybrid (Y2H) (protein-protein interaction verification)
Confirm the biological relevance of candidate variants to phenotypic traits with our precise validation platforms. Click the blue text link to learn more on the related page.
Sample Submission Guidelines
|
Sample Type |
Minimum Requirement |
Notes |
|---|---|---|
|
Genomic DNA |
>0.5 μg |
Concentration >20 ng/μL; OD260/280: 1.7-1.9; -20℃ storage, dry ice shipment |
|
Plant Tissue |
>0.5 g fresh weight |
Flash-frozen in liquid nitrogen; -80℃ storage and dry ice transport |
|
Animal Tissue |
>0.2 g fresh weight |
Flash-frozen in liquid nitrogen; -80℃ storage and dry ice transport |
|
Cells |
>1×106 cells |
Cell pellet, -80℃ storage, dry ice transport |
|
Blood (Anticoagulated) |
>1 mL |
Flash-frozen, -80℃ storage, dry ice transport |
| Sequencing Data* | Two parental and two progeny fastq files, plus reference genome fasta file | Direct BSA analysis available |
Notes:
· Parental lines should exhibit high homozygosity and clear phenotypic differences.
· Select at least 20 F2 individuals with extreme phenotypes for bulking.
· Species must have a reference genome available.
* Alternatively, clients can provide two parental and two progeny sequencing fastq files, along with the reference genome fasta file, for direct BSA analysis without the need for additional sequencing. This option offers a more cost-effective solution and significantly shortens the project timeline, with BSA analysis deliverables available within approximately 15 working days.
Service Details
· Trait Region Mapping: SNP-index and ED-based mapping to locate candidate regions.
· Candidate Gene Identification: Mining and annotation based on allele frequency shifts.
· Molecular Marker Development: Optional marker design for breeding applications.
· Functional Validation: qRT-PCR, EMSA, Y1H, and Y2H assays available based on client needs.
Frequently Asked Questions
Q1: How to construct bulks?
Select individuals with extreme phenotypes based on accurate phenotyping.
Q2: How to mix DNA for bulks?
Extract DNA separately from each individual, then mix in equal amounts to minimize background noise.
Q3: What makes suitable parental lines?
Parents should differ mainly in the target trait and have high homozygosity.
Q4: Can BSA analysis be performed without parental samples?
Yes. ED-based association mapping can be conducted using only two bulks.
Q5: Can BSA analyze multiple traits simultaneously?
No. Each BSA project focuses on a single trait.
Q6: Will BSA always locate a candidate region?
High probability if sample selection criteria are met. Region size varies depending on several factors.
Case Studies of BSA Analysis
Data Quality Control: High-quality sequencing data is fundamental. After obtaining raw data, we conduct comprehensive quality assessments, including base quality distribution and base composition analysis, to ensure data reliability.
SNP Detection: We detect single nucleotide polymorphisms (SNPs) caused by base substitutions using SNP-index and Euclidean Distance (ED) algorithms. SNP-index identifies significant differences in allele frequency between bulks, while ED measures distance-based differences to enhance mapping accuracy.
Indel Detection: Using GATK, we detect insertion-deletion (Indel) polymorphisms and integrate SNP and Indel association analyses to further narrow down candidate regions.
Variant Annotation and Functional Enrichment: Variants are annotated to prioritize genes causing stop gains/losses, nonsynonymous mutations, splice site alterations, or frameshifts. GO enrichment analysis highlights candidate genes involved in relevant biological pathways.

Experimental Setup
Sequencing Platform: Illumina Next-Generation Sequencing.
Analysis Tools: BWA (alignment), GATK (variant calling), SnpEff (variant annotation).
Algorithms: Combined use of SNP-index and ED for robust trait mapping.
Validation Techniques: qRT-PCR, EMSA, Y1H, Y2H.
Why Choose ProNet Biotech?
Rapid Turnaround: Accelerated project delivery without compromising quality.
Integrated Platform: Full pipeline from sequencing to functional verification.
Competitive Pricing: Cost-effective solutions tailored to your project scale.
Expert Support: Dedicated scientific consultants to guide your project to success.
References
[1] Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25(14):1754-60. doi: 10.1093/bioinformatics/btp324. Epub 2009 May 18. PMID: 19451168; PMCID: PMC2705234.
[2] McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010 Sep;20(9):1297-303. doi: 10.1101/gr.107524.110. Epub 2010 Jul 19. PMID: 20644199; PMCID: PMC2928508.
[3] Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012 Apr-Jun;6(2):80-92. doi: 10.4161/fly.19695. PMID: 22728672; PMCID: PMC3679285.
[4] Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol. 2012 Jan 22;30(2):174-8. doi: 10.1038/nbt.2095. PMID: 22267009.
[5] Hill JT, Demarest BL, Bisgrove BW, Gorsi B, Su YC, Yost HJ. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 2013 Apr;23(4):687-97. doi: 10.1101/gr.146936.112. Epub 2013 Jan 8. PMID: 23299975; PMCID: PMC3613585.
Ready to accelerate your trait mapping and gene validation projects?
Contact ProNet Biotech today to learn more about our BSA service solutions!
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry