Services
Providing Advanced Biological Technical Services for Researchers in Fundamental Science
Digital Protein/Nucleic Acid Interaction Prediction Model
AI in Biotechnology: Revolutionizing Protein and Nucleic Acid Interaction Analysis
Service Description
AI, as a revolutionary tool, has shown immense potential in the life sciences field. In the area of proteomics, machine learning and deep learning are widely applied to study protein interactions, protein-nucleic acid interactions, and protein-small molecule docking. ProNet Biotech's digital library screening technology leverages AI to predict protein interactions based on the three-dimensional structure of proteins, enabling efficient validation through experimental confirmation. This approach significantly reduces the time and cost associated with traditional experimental methods.
Application Areas
Our AI-driven services cater to various research areas in biotechnology, including:
Protein-Protein Interactions (PPI): Investigate disease mechanisms, signal transduction pathways, and predict protein functions. Enhance drug development and disease research with precise PPI predictions.
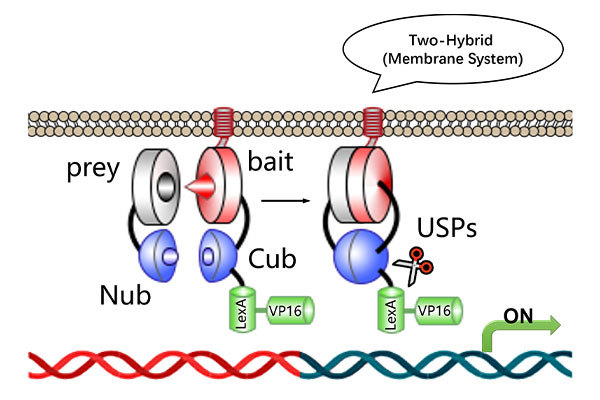
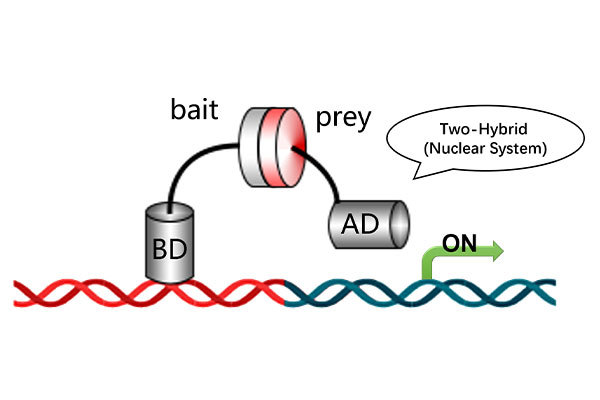
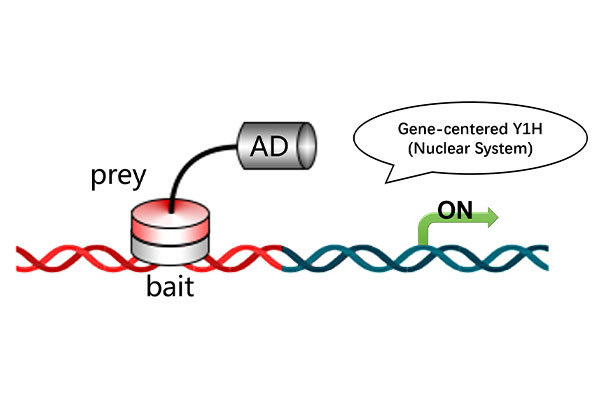
Protein-Nucleic Acid Docking: Explore the molecular interaction between proteins and nucleic acids, aiding in disease mechanism research, biomolecular complex structure prediction, and gene therapy development.
Virtual Sample Submission Standards
To get started with our AI-powered digital prediction models, we require the following sequences:
Protein-Protein Interaction | Protein-Nucleic Acid Docking |
Provide the CDS sequence or amino acid sequence of the protein. | Provide both the protein's CDS sequence or amino acid sequence, and the sequence of the nucleic acid. |
Traditional vs. AI-Driven Interaction Prediction
Traditional Methods:
Conventional protein-protein interaction (PPI) detection relies on experimental techniques like yeast two-hybrid (Y2H) and co-immunoprecipitation (Co-IP), which are time-consuming and have a low success rate for certain difficult-to-express proteins.
AI-Powered Prediction:
ProNet Biotech uses computational methods to predict potential interactions based on protein sequence or structural information, providing fast and accurate results with minimal experimental effort.
Service Process
ProNet Biotech's AI-based protein interaction prediction model follows a three-round screening process:
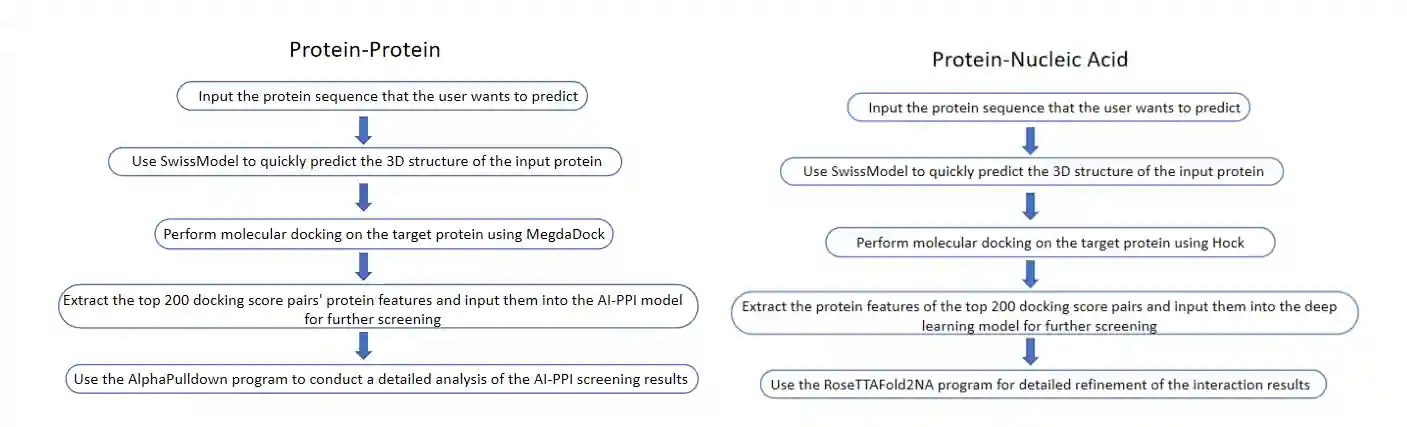
Protein interaction prediction model | ||
Service content | Working days | Delivery material |
1. Bait Gene Modeling Analysis | 1 | Complete Data Excel Sheet (including gene IDs, modeling scores, interaction status) |
2. Initial Screening Docking Assessment with DOCK | 2 | |
3. Deep Prediction Analysis of Large Model Interactions | 5 | |
4. Detailed Analysis | 5 | |
5. Data Organization and Analysis | 2 | |
Case Study: Protein-Protein Interaction Prediction
1. 3D Structural Modeling of the Bait Protein
The first step in protein-protein interaction prediction is constructing a 3D structural model of the bait protein. We use advanced AI algorithms to generate highly accurate models, with color coding to represent the confidence levels of various parts of the structure. Higher confidence indicates a more reliable model, providing a strong foundation for subsequent docking and interaction predictions.
2. Docking with Target Species Database
Next, we dock the bait protein model with the 3D structure database of the target species (digital library construction services available from ProNet Biotech). This step produces docking visualizations and docking scores to assess the potential interactions between the bait protein and other proteins in the target species.
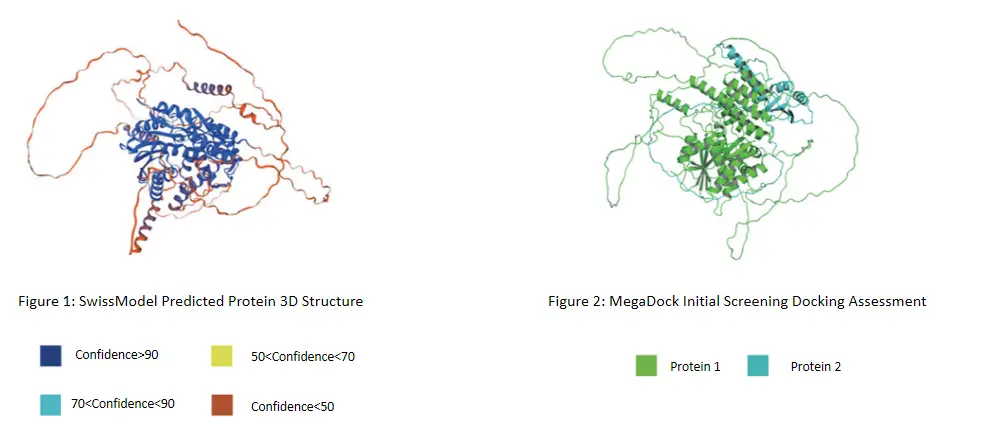
3. Feature Data Extraction:
Extract feature data of all docking proteins and input it into ProNet Biotech's AI-PPI model for detailed screening. This process generates protein annotations, interaction scores, and modeling scores.
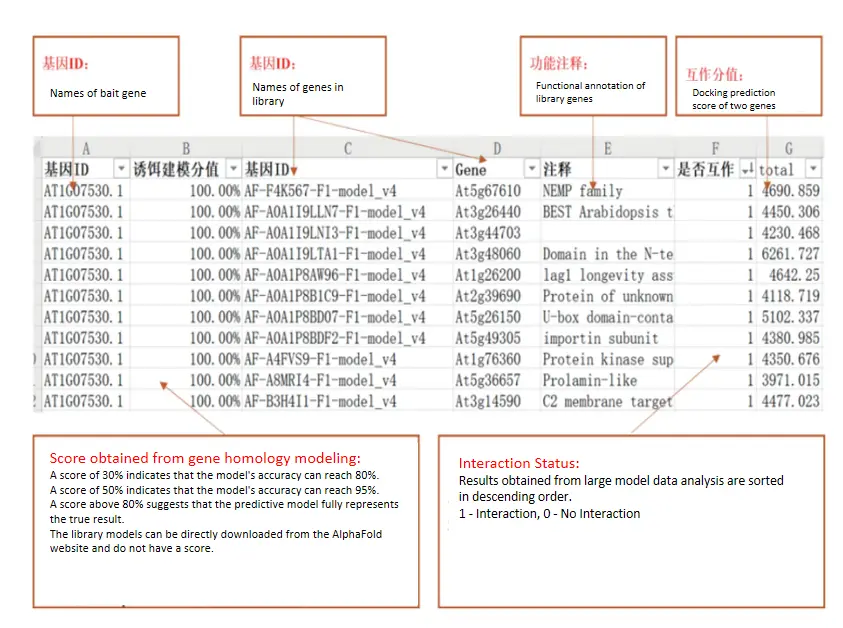
Figure 3. Deep Prediction Analysis of Large Model Interactions
4. Deep Prediction Analysis:
Use the AlphaPulldown program to perform in-depth analysis of the predicted interactions, focusing on potential false positives and the most promising interaction pairs. Confidence scores are visualized using color coding, where interactions from the STRING database are marked in red, and those from the Biogrid database are marked in yellow.
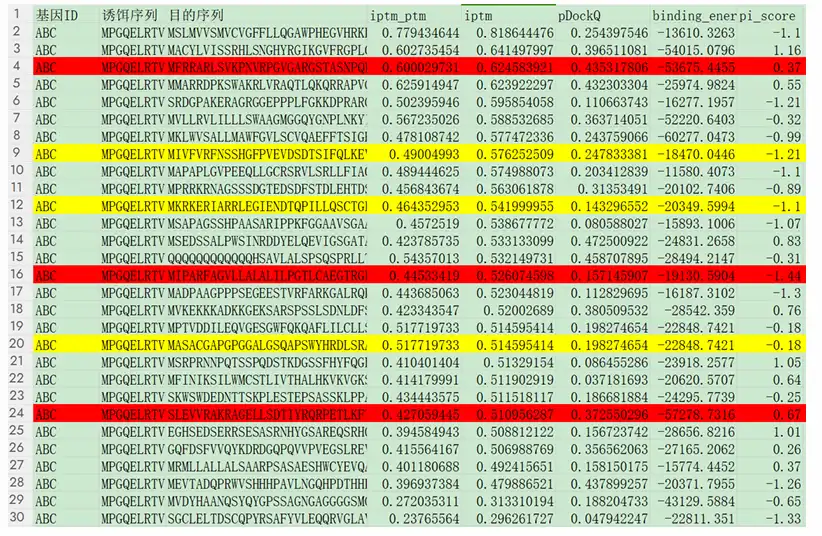
Figure 4. Interaction data, with interactions from the STRING database marked in red and those from the Biogrid database marked in yellow.
5. Interaction Interface Analysis:
Analyze the top protein pairs with the highest ipTM_pTM scores using PyMOL to visualize the interaction interface, including amino acids that form hydrogen bonds and the distances between them.
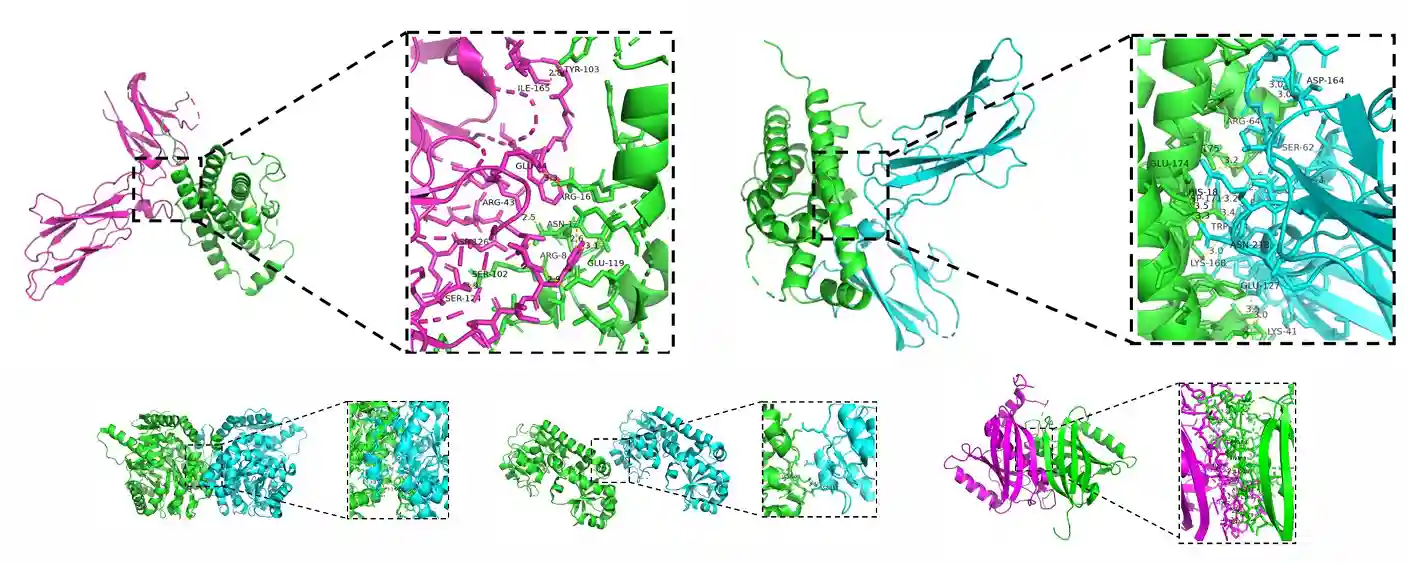
Figure 5. Interaction Interface Analysis
Why Choose ProNet Biotech?
Cutting-Edge AI Technology: Our AI models are developed in-house and constantly updated to stay at the forefront of research in protein interaction prediction.
Streamlined Process: The three-round screening process is designed to deliver accurate results quickly, reducing time and cost for researchers.
Comprehensive Support: We offer detailed analysis and visualizations to help researchers make informed decisions based on our predictions.
Research-Grade Accuracy: Our models are tailored for high-precision, research-driven applications in biotechnology, making them ideal for use in drug discovery, gene therapy, and more.
Contact Us
For more information about how our AI-driven protein interaction prediction models can support your research, or to submit your samples, please [Contact Us] today!
Disclaimer: All products and services provided by ProNet Biotech are for research purposes only and may not be used for diagnostic or therapeutic purposes.
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry

Contact Us

Tel: +86 025-85205672

Email: info@pronetbio.com

Address: Building 3C, Nanjing Xianlin Zhigu,
Qixia District, Nanjing, Jiangsu, China, 210033

Need more info?
Let's connect!