Services
Providing Advanced Biological Technical Services for Researchers in Fundamental Science
AI-Based Protein Interaction and Binding Site Prediction Analysis
AI-Powered Protein Interaction Prediction and Binding Site Analysis for Advanced Molecular Research
Product Description
ProNet Biotech offers cutting-edge AI-powered solutions for protein interaction prediction and binding site analysis. Our intelligent system evaluates the confidence of protein-protein and protein-nucleic acid interactions, providing reliable confidence scores that can guide your research and subsequent experiments.
Whether you're exploring protein-antibody affinity, protein-protein interactions, or protein-nucleic acid binding, our AI-driven service eliminates the need for expensive reagents and long experimental waits, offering you faster and more reliable results.
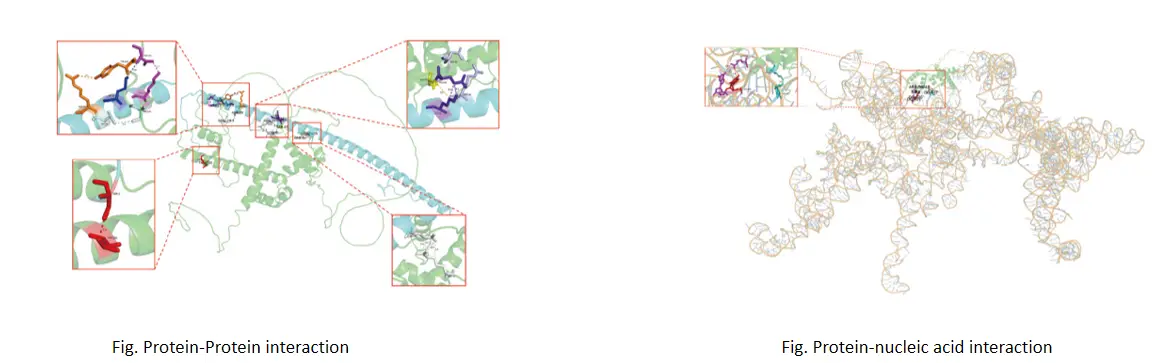
Technical Advantages
Zero Experimental Costs: Fully automated service eliminates the need for experimental operations and costly reagents.
Rapid Candidate Gene Screening: Quickly generate protein interaction maps, reducing the waiting time typically associated with experimental setups.
Reliable Confidence Analysis: Our system provides confidence scores to narrow down the pool of interacting proteins or genes, enabling you to focus only on the most promising candidates.
Journal-Compliant High-Resolution Images: Receive 3D protein structure analysis images that meet the formatting requirements of a variety of academic journals.
Worry-Free Follow-Up Experiments: We offer an optional yeast one-on-one validation service to further confirm your screening results, providing accurate quantitative analysis and enhancing the reliability of your findings.
Application Scenarios
Protein-Antibody Affinity Prediction: Predict the interaction strength between proteins and antibodies for antibody development.
Protein-Protein Interaction Modeling: Generate 3D interaction models to analyze protein-protein interactions and binding sites.
Protein-Peptide Binding Affinity: Evaluate the binding strength between proteins and peptides.
Enzyme-Substrate Binding Affinity: Predict the interaction between enzymes and their protein substrates.
Three-Protein Interaction Analysis: Analyze interactions involving three proteins for more complex interaction studies.
Protein-Nucleic Acid Interaction Analysis: Study the interactions between proteins and DNA or RNA to support molecular biology research.
Service Content
|
Service |
Working Days |
Deliverables |
|---|---|---|
|
Protein Interaction Confidence Analysis |
5 days |
Electronic analysis results |
|
Downstream Experiments (Optional) |
Varies |
Yeast one-on-one validation report |
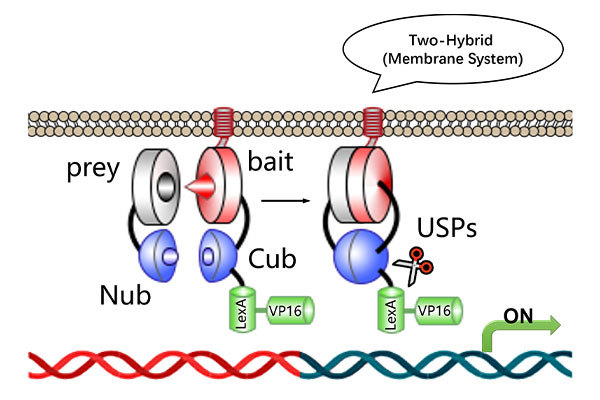
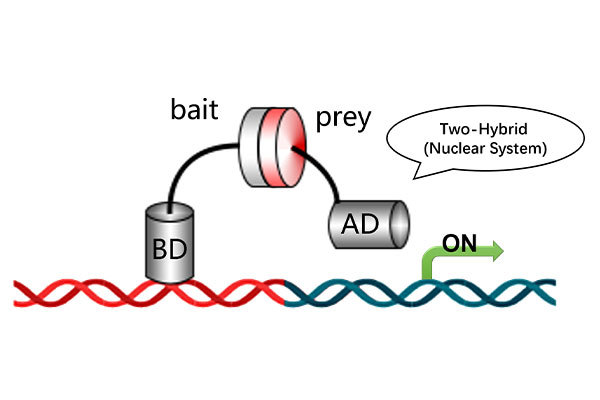
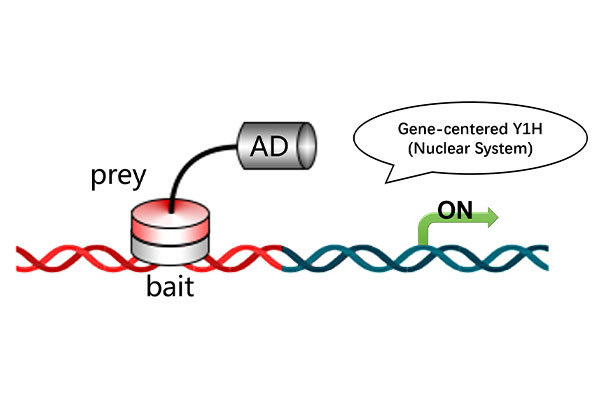
Downstream Experiments for Protein-Protein Interaction and Binding Site Analysis
Identify potential binding site amino acid sequences based on the nucleic acid sequence of the coding protein.
Mutate each site to alanine and perform yeast one-on-one verification to confirm binding site interactions.
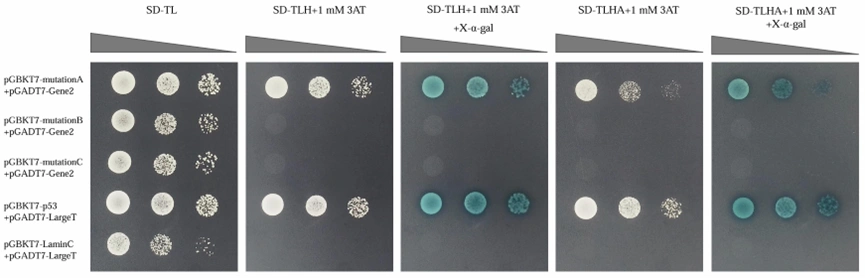
Figure 1. Yeast one-on-one verification of protein binding sites.
References
· Yan Y, Tao H, He J, Huang S-Y. The HDOCK server for integrated protein-protein docking. Nature Protocols, 2020; doi: https://doi.org/10.1038/s41596-020-0312-x.
· Yan Y, Zhang D, Zhou P, Li B, Huang S-Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365-W373.
· Yan Y, Wen Z, Wang X, Huang S-Y. Addressing recent docking challenges: A hybrid strategy to integrate template-based and free protein-protein docking. Proteins 2017;85:497-512.
· Huang S-Y, Zou X. A knowledge-based scoring function for protein-RNA interactions derived from a statistical mechanics-based iterative method. Nucleic Acids Res. 2014;42:e55.
· Masahito Ohue, Takehiro Shimoda, Shuji Suzuki, Yuri Matsuzaki, Takashi Ishida, Yutaka Akiyama. MEGADOCK 4.0: an ultra-high-performance protein-protein docking software for heterogeneous supercomputers, Bioinformatics, 30(22): 3281-3283, 2014.
· Takehiro Shimoda, Shuji Suzuki, Masahito Ohue, Takashi Ishida, Yutaka Akiyama. Protein-Protein Docking on Hardware Accelerators: Comparison of GPU and MIC Architectures, BMC Systems Biology, 9(Suppl 1): S6, 2015.
Contact Us Today for a Free Consultation!
Maximize your research efficiency with ProNet Biotech's AI-based protein interaction prediction and binding site analysis. Request a custom analysis and get started on your next groundbreaking project.
Disclaimer: All products and services provided by ProNet Biotech are for research purposes only and may not be used for diagnostic or therapeutic purposes.
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry

Contact Us

Tel: +86 025-85205672

Email: info@pronetbio.com

Address: Building 3C, Nanjing Xianlin Zhigu,
Qixia District, Nanjing, Jiangsu, China, 210033

Need more info?
Let's connect!