Yeast Two-Hybrid Screening Service (Membrane Protein)
Membrane Yeast Two-Hybrid (MYTH) Screening Service | Protein Interaction Analysis & Network Mapping
Professional Membrane Protein Interaction Analysis Service
Our Membrane Yeast Two-Hybrid (MYTH) screening service (Service Code: SRY8010) is designed to membrane protein-protein interaction analysis using membrane protein cDNA libraries. Our cutting-edge system specializes in analyzing interactions between membrane proteins and cytoplasmic proteins, offering unparalleled sensitivity and reliability in protein interaction studies.
We offer a comprehensive solution for analyzing protein-protein interactions, combining traditional Y2H screening with next-generation sequencing technology.
Why Choose Our Advanced MYTH Screening Service?
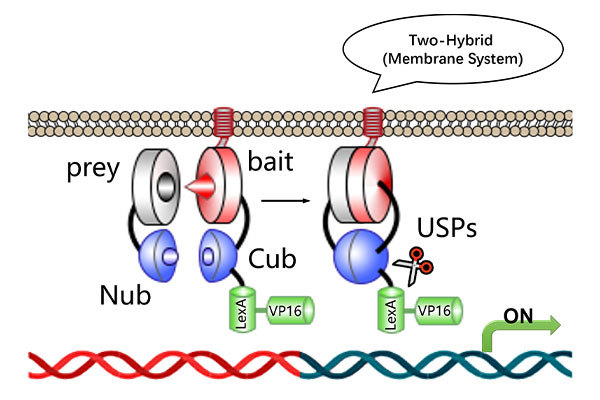
Our Membrane Yeast Two-Hybrid (MYTH) screening service combines industry-leading accuracy with advanced split-ubiquitin technology to deliver reliable, high-sensitivity protein-protein interaction analysis. Key advantages include:
High Accuracy & Sensitivity – Detect protein-protein interactions in vivo without nuclear localization signals, using optimized NubG/Cub domains to minimize steric hindrance and reduce false positives.
Wide Application Scope – Suitable for membrane proteins in plasma membrane, ER, mitochondria, Golgi, as well as cytoplasmic and organelle proteins.
Comprehensive Insights – Functional annotations, interaction strength analysis, and pathway mapping (GO, KEGG) for deeper biological interpretation.
Fast Turnaround & Competitive Pricing – Optimized workflows ensure up to 50 working days delivery with cost-effective solutions for global clients.
Our Advanced Technical Platform
Powered by a refined split-ubiquitin MYTH system, our platform is engineered for precision and reproducibility:
In Vivo Interaction Detection – Directly monitor protein interactions inside living yeast cells.
Transcription-Independent Activation – UBP-mediated LexA cleavage enables detection even when bait proteins contain activation/repression domains.
High-Stringency Screening – Multi-reporter system (His, Ade, LacZ) with diverse promoter regions reduces background noise and improves true positive rate.
Professional Quality Assurance
Every MYTH screening project follows rigorous quality control procedures to ensure reproducible, publication-ready results:
· Standardized experimental protocols and validated workflows.
· Multiple checkpoints including autoactivation tests, re-screening, and independent verification.
· Detailed, high-resolution reporting with raw data, analysis files, and visual interaction networks.
· Dedicated technical support from project initiation to post-delivery consultation.
MYTH Screening Timeline and Process Flow
Our streamlined workflow ensures high-throughput and reliable membrane protein interaction analysis within an optimized schedule. Below is the detailed project timeline for our Membrane Yeast Two-Hybrid (MYTH) Screening Service:
Phase | Workflow Step | Working Days | Description |
|---|---|---|---|
Phase 1 | Transmembrane Structure Analysis | < 1 Day | Comprehensive transmembrane structure analysis and comprehensive project planning. |
Phase 2 | Gene Synthesis & Vector Construction | 10 Days | Codon-optimized gene synthesis and cloning into bait/prey vectors for Y2H system. |
Phase 3 | Autoactivation Test of Bait Plasmid | 10 Days | Self-activation testing to eliminate false positives and optimize bait design. |
Phase 4 | Co-Transformation, Yeast Screening & Sanger Sequencing | 15 Days | Yeast co-transformation, high-stringency selection using MYTH system (20 plates in total), and sequencing of 96 positive clones. |
Phase 5 | Positive Clone Re-Screening & Temporary Data Report | 5 Days | Retransformation validation of positive clones after Sanger sequencing on SD-TL, SD-TLH, SD-TLH + X-α-gal, SD-TLHA, and SD-TLHA + X-α-gal plates and preliminary results summary. |
Phase 6 | Next-Generation Sequencing & Final Report | 10 Days | Collection of all rest clones for NGS, bioinformatics analysis, interaction network visualization, and strength analysis. |
Total Duration: ≤ 50 Working Days
Service Deliverables
· Positive clone sequences (≥15 sequences based on Sanger sequencing and re-screening)
· Bait plasmid (optional)
· Comprehensive NGS data and bioinformatics analysis
· Detailed functional annotations (GO, KEGG) with interaction strength analysis
· Professional research report including:
· Complete methodology
· Step-by-step experimental procedures
· High-resolution images
· Expert data interpretation
Featured Client Publications
One of our clients has recently published a research article in Forestry Research acknowledging ProNet Biotech Co., Ltd. This study successfully applied our Yeast Two-Hybrid (Y2H) screening platform to identify interacting proteins of Malus sieversii ABA receptor PYL8, followed by point-to-point validation and BiFC assays.
Citation:
Identification of Malus sieversii ABA receptor PYL8 interacting proteome using Y2H-seq
Forestry Research (2025), 5: e012
https://www.maxapress.com/article/doi/10.48130/forres-0025-0012
doi: 10.48130/forres-0025-0012
Ready to Advance Your Research?
Contact us today for a case deliverables report, detailed consultation and service quotation. Let us help you unlock new insights in protein interaction research. When inquiring about this Membrane Yeast Two-Hybrid (MYTH) screening service, please reference Service Code: SRY8010.
Y2H Screening FAQs
1. Should chloroplast-localized proteins be screened using the nuclear or membrane system?
This depends on whether the protein has a transmembrane domain. For proteins with transmembrane regions, membrane Y2H screening is preferred. For soluble or non-membrane-targeted proteins, the nuclear system may be more appropriate.
2. How does the membrane Y2H screening process work?
You provide your bait gene, and we clone it into a pBT3-series vector. After autoactivation testing, we co-transform with the prey library. We sequence 96 positive clones and perform retransformation validation. Remaining clones from large screening plates are scraped and subjected to high-throughput sequencing (NGS) for comprehensive analysis.
3. Are the proteins screened using a membrane library in yeast two-hybrid experiments always membrane proteins?
Our advanced MYTH system can detect various protein types, including membrane, nuclear, and organelle proteins. The diversity depends on the specific cDNA library used in the screening process. Positive clones are identified based on interaction, not subcellular localization.
4. How to choose the right vector in membrane Y2H screening?
Vector selection depends on the orientation of the bait protein’s N- and C-termini. Proper positioning ensures effective recognition and cleavage by ubiquitin-specific proteases (UBPs), activating the reporter gene. Different bait protein topologies require tailored vector choices (e.g., pBT3-N, pBT3-STE, or pBT3-SUC).
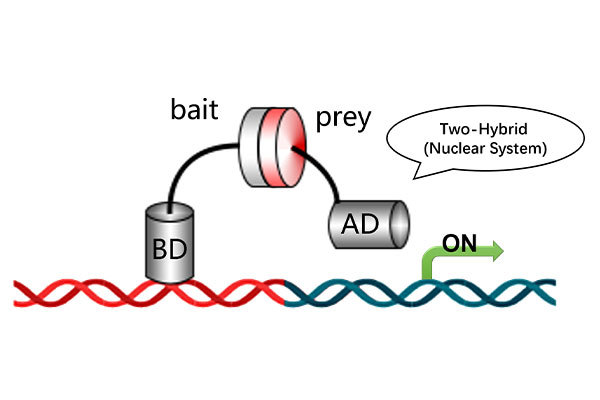
5. What are the key differences between nuclear and membrane Y2H systems?
· Yeast strains: The nuclear system uses Y2HGold; the membrane system uses NMY51.
· Library transformation: The nuclear system supports both co-transformation and mating; the membrane system uses co-transformation only.
· Screening mechanism: The nuclear system relies on Gal4-based activation, while the membrane system employs split-ubiquitin and LexA-based transcription.
6. Should the signal peptide be removed when synthesizing membrane protein genes?
Yes. Signal peptides can affect proper membrane localization and fusion protein formation. Removal ensures accurate bait protein behavior in yeast.
7. What if both ends of the bait protein are extracellular?
In this case, the bait gene should be truncated to expose an intracellular domain. This allows proper vector fusion and reporter activation in the membrane Y2H system.
8. Why is functional validation important before membrane system screening?
Functional validation ensures that the Bait-Cub fusion protein is correctly localized and functional in yeast cells. Using a control vector (pOst1-NubI), we verify bait expression to avoid false positives.
☆ Related Optional Service: Membrane Yeast Two-Hybrid One-to-One Interaction Validation
Y2H Validation Service Overview
Our Membrane Yeast Two-Hybrid One-to-One Interaction Validation service is designed for precise analysis of specific protein-protein interactions within membrane systems. This service is ideal for researchers aiming to validate screening results or investigate specific protein-protein interactions, ensuring data accuracy and expanding research possibilities.
Y2H Validation Experimental Workflow
Phase 1: Transmembrane Analysis (< 1 Day)
Transmembrane structure analysis.
Phase 2: Construction Phase (10 Days)
Gene synthesis (codon optimization).
Vector construction: Bait protein A vector constructed to pBT3-N/SUC/STE; protein B vector constructed to pPR3-N.
Phase 3: Transformation (7 Days)
Recombinant plasmids transformed into yeast cells and autoactivation test performed.
Phase 4: Functional Validation (3 Days)
Bait plasmid + pOst1-NubI used for validation.
Phase 5: Interaction Validation (10 Days)
Set up positive and negative control groups for interaction validation.
Y2H Validation Deliverables
· Recombinant plasmids (Optional).
· A digital experiment report.
Statement: Our services are exclusively for research applications and are not intended for diagnostic or therapeutic use.
Related Product: Yeast Two-Hybrid (Membrane System) Vector Kit [CAT:RY8010]
Related Services: Explore our Co-IP and Pull-Down Assays for advanced protein-protein interaction validation.
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry