Services
Providing Advanced Biological Technical Services for Researchers in Fundamental Science
Luciferase Complementation Assay (LCA)
Service Overview
The Luciferase Complementation Assay (LCA) is an advanced, highly sensitive method designed for studying protein-protein interactions. In this assay, luciferase is split into two fragments: the N-terminal fragment (NLuc) and the C-terminal fragment (CLuc). Each fragment is fused to a different target protein. When these target proteins interact, the luciferase fragments come together to reconstitute a functional luciferase enzyme, which emits a luminescent signal. This luminescence can then be used to assess and quantify protein interactions.
LCA is widely used in molecular biology, drug discovery, and biological research to provide valuable insights into protein dynamics and interactions, contributing to the development of therapeutic strategies and enhancing understanding of complex biological systems.
Key Benefits of LCA
High Sensitivity: Detect and quantify even weak protein-protein interactions, making it suitable for both qualitative and quantitative analysis.
Quantitative & Qualitative Results: Obtain both qualitative and quantitative results from a single experiment.
Reliable Controls: Positive and negative controls ensure reliable experimental processes and validation.
Complementary Verification: Multiple protein interaction assays can be used to confirm results, increasing reliability and accuracy.
Applications of LCA
Protein-Protein Interaction Studies: LCA is a powerful tool for studying complex interactions between proteins in living cells, providing a deeper understanding of molecular functions.
Drug Discovery & High-Throughput Screening: Ideal for identifying small molecules or compounds that can modulate protein interactions, advancing the drug discovery process.
Molecular Pathway Analysis: Use LCA to explore signaling pathways, protein complexes, and cellular mechanisms in various biological processes.
How LCA Works
In LCA, luciferase is split into two non-functional fragments (NLuc and CLuc), each fused to a target protein. When the two target proteins interact, the luciferase fragments come together, restoring luciferase activity and generating a luminescent signal. The resulting signal can be measured using either a plate reader or in vivo imaging, allowing for real-time observation of protein interactions.
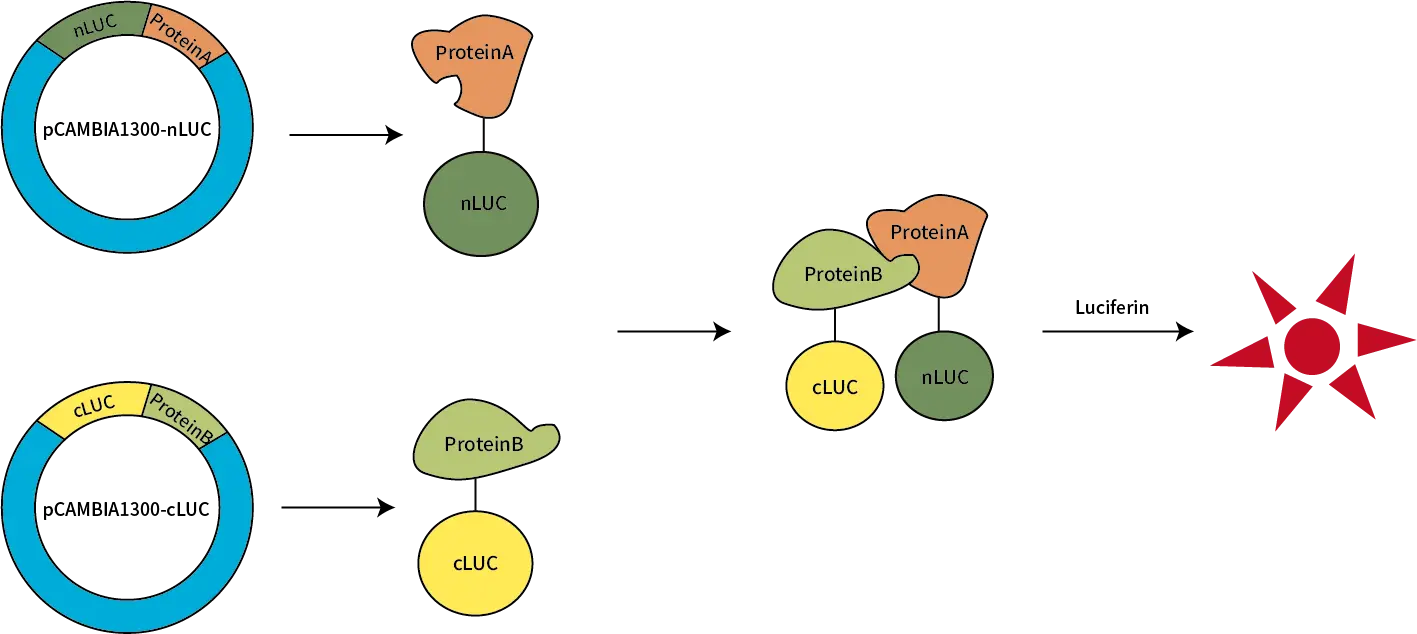
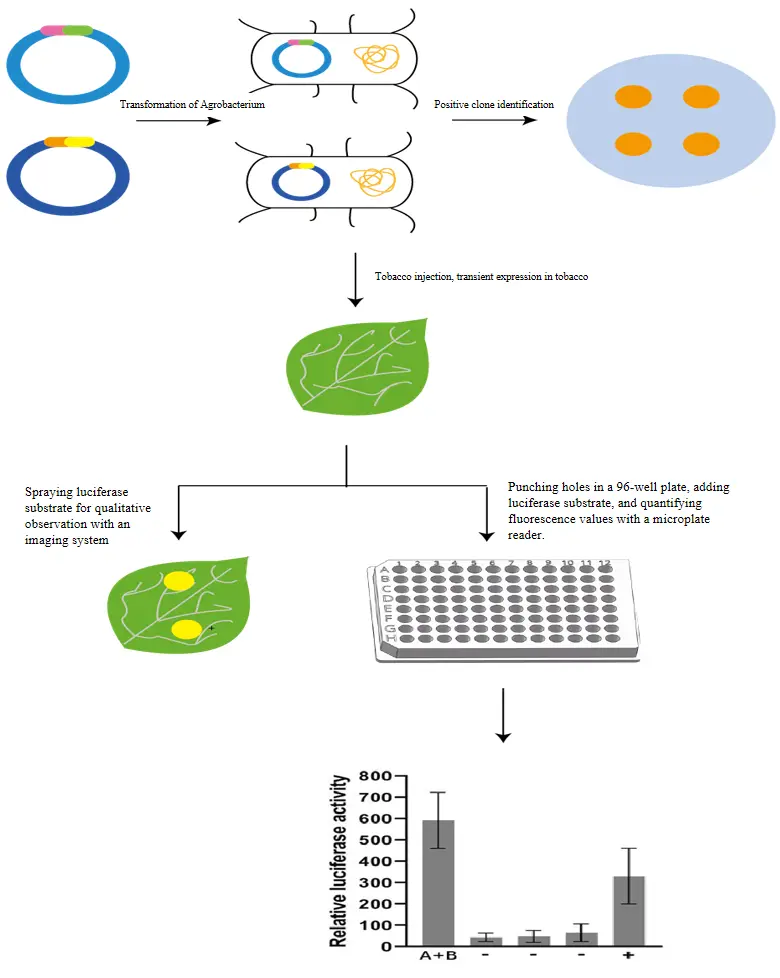
Service Workflow & Requirements
Sample Submission: Please provide the CDS sequences for the two target proteins to be tested.
Service process | Working days | Delivery material |
Customer place an order | 10 | 1. Plasmid and sequencing verification report for gene synthesis/vector construction 2. Raw data and result images 3. Standard experimental report (including experimental steps and procedures) |
Confirmation of experimental materials | ||
Gene synthesis/vector construction | ||
Tobacco injection | 10 | |
Luciferase activity observation | ||
Quantitative analysis | 5 | |
Data organization | ||
Total | 25 |
Case Study
To examine whether proteins A and B interact, the assay was performed using both in vivo imaging and plate reader techniques for qualitative and quantitative analysis. As shown in Figure 1A, the in vivo imaging reveals strong luminescence in both experimental groups, indicating a positive interaction between proteins A and B, even stronger than the positive control. Quantitative analysis from the plate reader (Figure 1B) confirms that the luciferase activity in the experimental groups significantly exceeds that of the negative controls, supporting the clear interaction between proteins A and B.
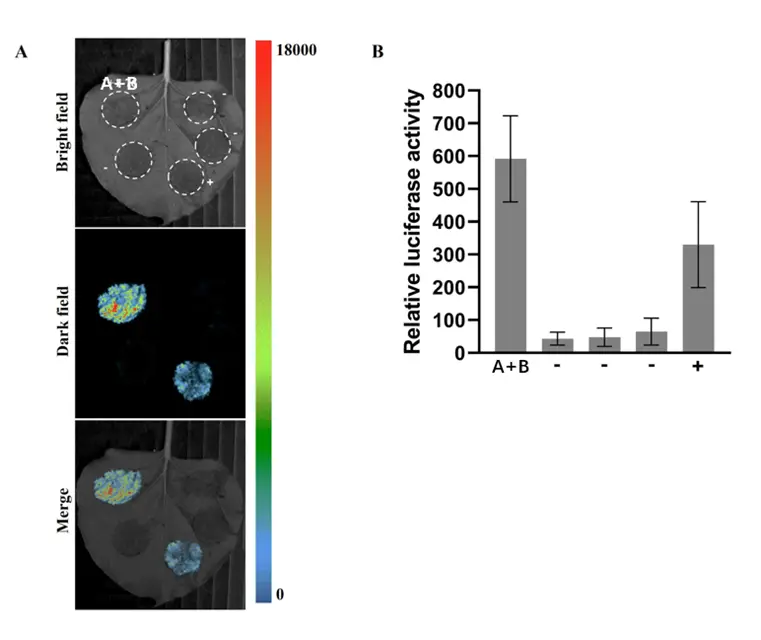
Figure 1: Protein Interaction Results
A. In Vivo Imaging: Visualization of protein interactions between A and B.
B. Plate Reader: Quantitative analysis of luciferase activity for A-B interaction.
Frequently Asked Questions (FAQs)
Q1: What should I do if the luminescence signal is weak?
Verify vector construction and protein expression.
Increase transfection efficiency and optimize substrate concentration.
Q2: How can I improve experimental accuracy?
Use multiple replicates to ensure statistical relevance.
Inject controls and experimental groups on the same leaf to standardize growth conditions.
Ensure consistent background by injecting 3-5 leaves per condition.
Adjust washing conditions to minimize nonspecific binding, and use appropriate blocking reagents.
Avoid endogenous luciferase leakage by reducing cell lysis.
Q3: How long does it take to get results from the LCA service?
The typical turnaround time is 25 working days, including plasmid construction, experimental analysis, and data processing.
References
Zhu F, Wang K, Li D, Liu Z, Li M, Wang Z, Li X, Lan X, Guan Q. OsSAP6 Positively Regulates Soda Saline-Alkaline Stress Tolerance in Rice. Rice (N Y). 2022 Dec 27;15(1):69. doi: 10.1186/s12284-022-00616-x. PMID: 36574073; PMCID: PMC9794665.
Yu H, Wang Q, Zhang Z, et al. Genetic Mapping of the Gmpgl3 Mutant Reveals the Function of GmTic110a in Soybean Chloroplast Development. Front Plant Sci. 2022 May 26;13:892077. doi: 10.3389/fpls.2022.892077. PMID: 35693168; PMCID: PMC9178232.
Disclaimer: Our products are intended for research purposes only and are not for diagnostic or therapeutic use.
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry

Contact Us

Tel: +86 025-85205672

Email: info@pronetbio.com

Address: Building 3C, Nanjing Xianlin Zhigu,
Qixia District, Nanjing, Jiangsu, China, 210033

Need more info?
Let's connect!