Services
Providing Advanced Biological Technical Services for Researchers in Fundamental Science
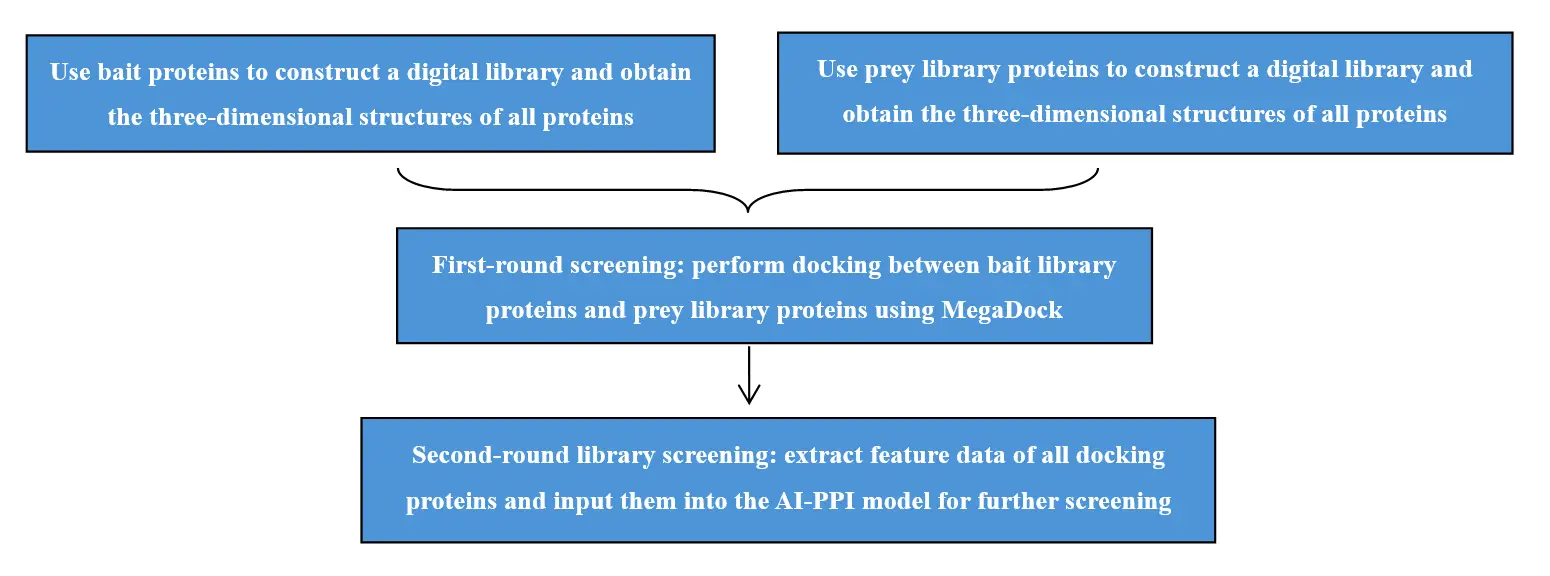
AI-Powered Digital Library-to-Library Interaction Screening
High-Throughput Digital Library-to-Library Interaction Screening Using AI-PPI and MEGADOCK
Interaction screening experiments often focus on a specific bait gene to perform a “one-to-many” screening against numerous candidate genes in a library, thereby obtaining regulatory information centered on that bait gene. In contrast, library-to-library screening constructs two libraries (A: bait library; B: prey library) and performs a “many-to-many” screening between them to reveal interaction network information of genes from both libraries.
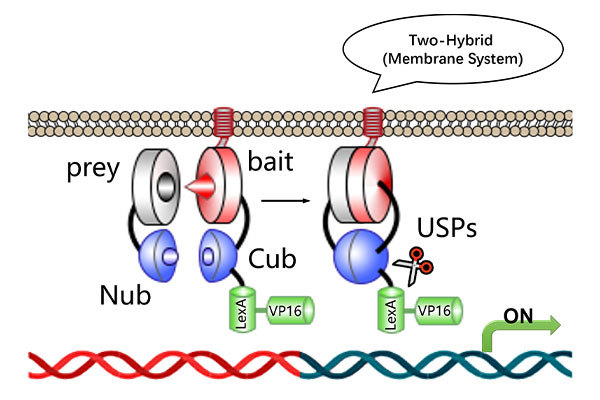
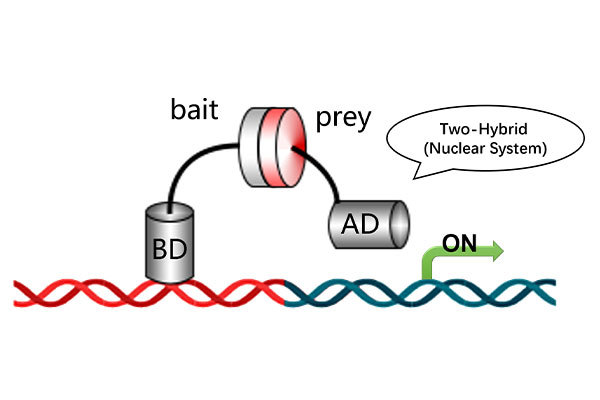
The traditional library screening technology is CrY2H-seq, established in 2017, which is based on the yeast two-hybrid principle and conducted within a yeast system. Nowadays, with the advancement of artificial intelligence and deep learning, it is possible to digitally construct two libraries using computational deep learning methods and predict protein-protein interactions between these two digital libraries through computational analysis.
Technical Advantages
1. Saves library construction steps: Provides ready-to-use whole-genome digital libraries for over 100 species and transcription factor libraries for more than 20 species, enabling immediate screening.
2. Two-stage screening strategy: Uses MegaDock for initial coarse screening and AI-PPI for precise secondary screening and deep analysis, accurately narrowing down candidate interaction pairs.
3. Cross-species interaction prediction: Applicable to studying cross-species biological interactions such as virus-host interactions.
4. High-quality visuals: Downstream AOS analysis offers protein 3D models (overview + close-up) combined with binding site analysis, clearly displaying binding sites, hydrogen bonds, and distance information to provide comprehensive interaction details.
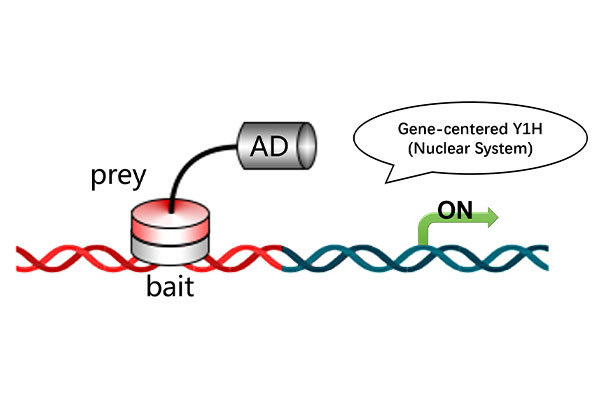
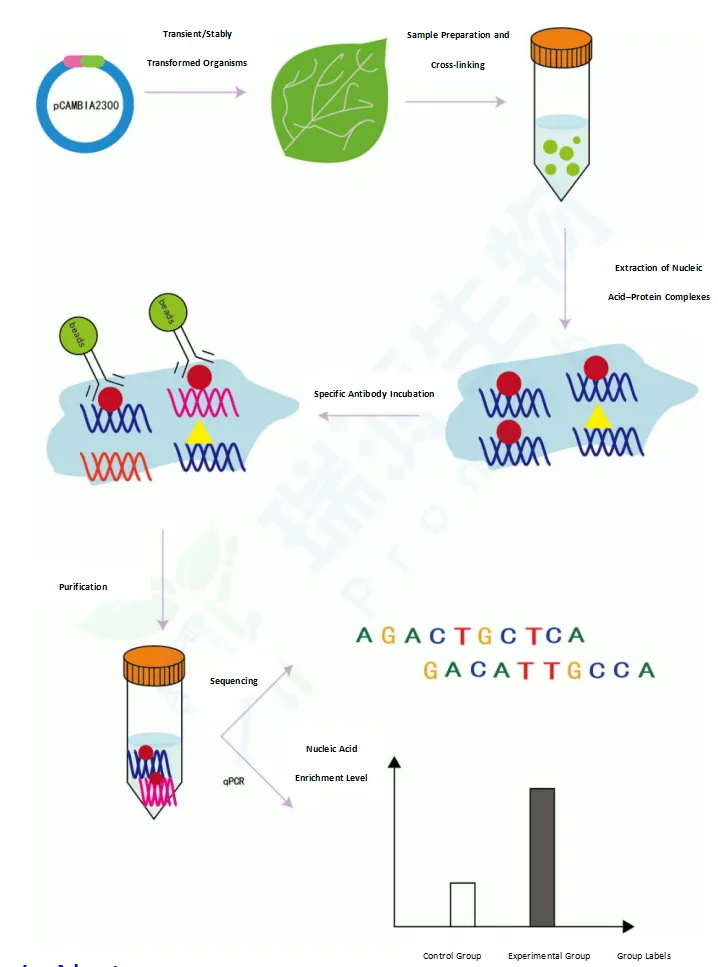
5. Complete downstream validation system: Own platform supports various validation experiments including yeast two-hybrid, yeast one-hybrid, CoIP, EMSA, BiFC, and dual-luciferase assays.
Service Process
Sample Submission Requirements
Sample Type | Requirement | Remark |
|---|---|---|
Bait library | CDS file required | If a specific database version is required, please specify clearly |
Prey library |
Service Process, Timeline, and Delivery
Order placement | Working day | Delivery materials |
Library gene modeling and analysis | Total number of pairs from multiplying two libraries (X): 10,000 < X ≤ 500,000, 7 working days 500,000 < X ≤ 1,000,000, 15 working days 1,000,000 < X ≤ 2,000,000, 20 working days 2,000,000 < X ≤ 4,000,000, 25 working days 4,000,000 < X ≤ 6,000,000, 40 working days 6,000,000 < X ≤ 8,000,000, 50 working days 8,000,000 < X ≤ 10,000,000, 65 working days | Complete Excel file of two-round digital library screening (including gene IDs, modeling scores, and interaction status) |
| DOCK preliminary docking evaluation | ||
| Deep interaction prediction analysis using large models | ||
| Data organization and analysis |
Frequently Asked Questions (FAQ)
Q1: Can the digital screening results be 100% experimentally validated?
The advantage of digital screening lies in its short turnaround time and high cost-effectiveness, making it a highly efficient initial screening approach. The docking results are based on AI deep learning, so false negatives and false positives are inevitable. Therefore, subsequent experimental validation is necessary to confirm positive interactions. According to existing case studies, the prediction accuracy of this method can exceed 60%.
Q2: How does this method compare to traditional yeast two-hybrid (Y2H) library screening?
Dimension | Digital Library-to-Library Screening | Yeast Library-to-Library Screening |
|---|---|---|
Principle | Computational models simulate structure/sequence binding | Experimental yeast growth reporter system |
Required Materials | Protein sequences or structural models | Constructed bait and prey plasmid libraries |
Throughput & Efficiency | Extremely high; millions of interactions in days | Low throughput; weeks to months required |
Accuracy & False Positives | Algorithm- and data-dependent; false positives possible | High accuracy but may suffer from biological background noise |
Case Report
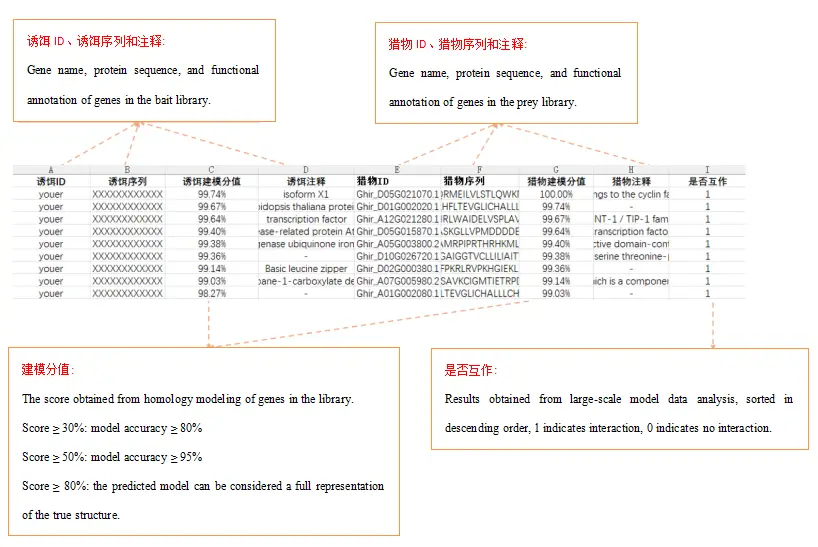
Screening results
Total: interaction score—the predicted docking score between two genes; the higher the score, the greater the likelihood of interaction;
Interaction status: results obtained from large-scale model data analysis, sorted in descending order, 1 indicates interaction, 0 indicates no interaction.
References
Ohue M, Matsuzaki Y, Uchikoga N, et al. MEGADOCK: an all-to-all protein-protein interaction prediction system using tertiary structure data[J]. Protein peptide letters, 2014, 21(8):766-778.
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry

Contact Us

Tel: +86 025-85205672

Email: info@pronetbio.com

Address: Building 3C, Nanjing Xianlin Zhigu,
Qixia District, Nanjing, Jiangsu, China, 210033

Need more info?
Let's connect!