Subcellular Localization Services
Comprehensive Subcellular Localization Services for Plant and Mammalian Systems
▼ Subcellular Localization Service Overview
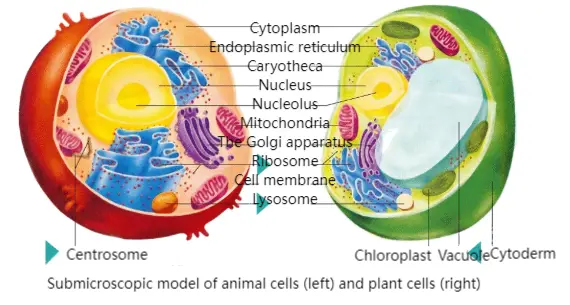
Subcellular localization refers to the specific location of biological macromolecules such as proteins, peptides, or lipids within a cell. After translation in the cytoplasm, proteins are directed by signal peptides to their functional destinations such as the nucleus, mitochondria, endoplasmic reticulum, or cell membrane. This intracellular targeting is critical, as correct localization is essential for proper protein function, metabolism, and molecular interactions. Mislocalization may severely impact cell function and even organismal survival. Therefore, understanding subcellular localization is a key step in functional genomics, molecular biology, and biomedical research.
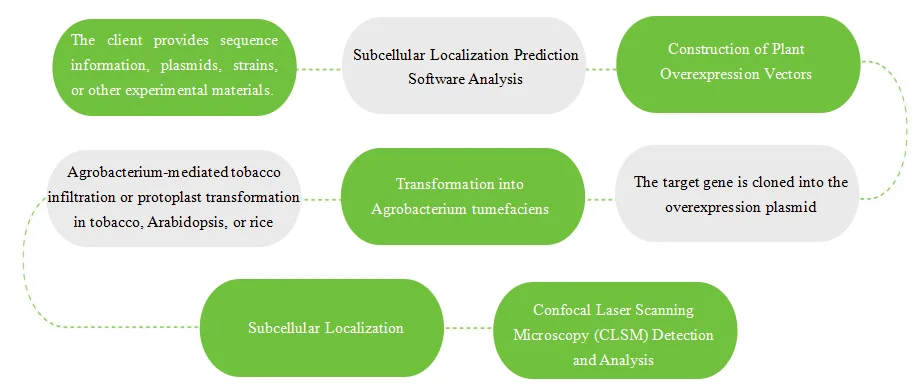
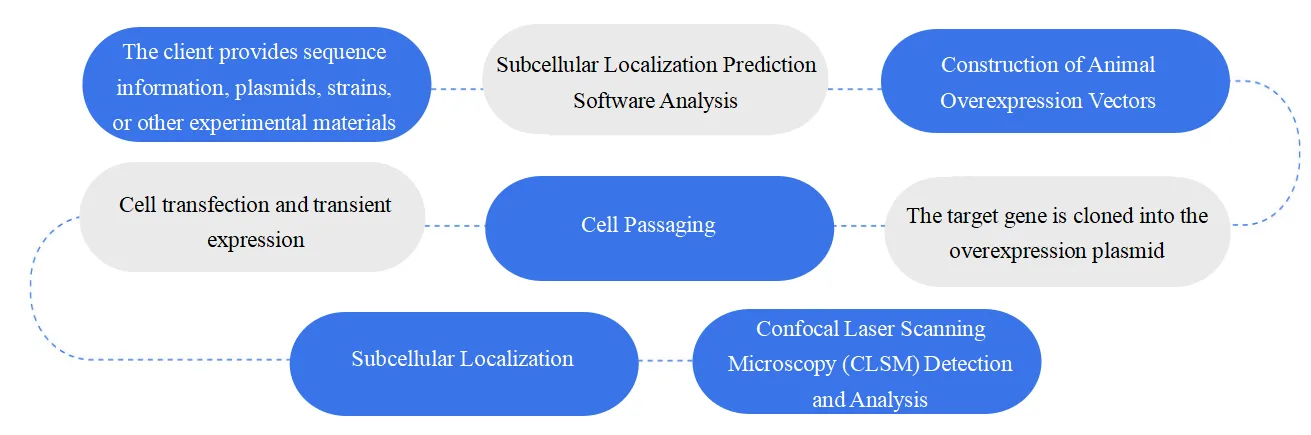
Service Workflow
Plant Subcellular Localization:
Animal Subcellular Localization:
Co-localization Markers:
List of Common Subcellular Markers for Localization in Animal Cells
Organelle | Marker Protein | Reference |
Nucleus | LEF1 | Chaturvedi NK et al., 2014 |
Plasma Membrane | STOML3 | Chaturvedi NK et al., 2014 |
Mitochondria | MRPL15 | Chaturvedi NK et al., 2014 |
Lysosome | LGMN | Chaturvedi NK et al., 2014 |
Endoplasmic Reticulum (ER) | ASPH | Chaturvedi NK et al., 2014 |
Golgi Apparatus | HS2ST1 | Chaturvedi NK et al., 2014 |
Peroxisome | ZADH2 | Chaturvedi NK et al., 2014 |
List of Common Subcellular Markers for Localization in Plant Cells
Organelle | Marker Protein | Reference |
Endoplasmic Reticulum | AtWAK2 | Nelson BK et al., 2007 |
Golgi Apparatus | AtXyIT | Pagny S et al., 2003 |
Vacuolar Membrane | At2g36830 | Nelson BK et al., 2007 |
Peroxisome | PTS1 | Nelson BK et al., 2007 |
Mitochondria | ScCoX4 | Nelson BK et al., 2007 |
Plastid (Chloroplast) | LOC107766567 | Nelson BK et al., 2007 |
Plasma Membrane | AtPIP2a | Nelson BK et al., 2007 |
Nucleus | NbH2b | Romit Chakrabarty et al., 2007 |
Technical Advantages
Mature Technical Approach: We offer subcellular localization and co-localization services based on a high-precision subcellular co-localization analysis platform, efficiently supporting research on protein-protein interaction mechanisms and gene function.
Comprehensive Service System: Using transformation systems including tobacco leaves, protoplasts (tobacco, Arabidopsis, rice), and mammalian 293T cells, we provide customized technical services ranging from plasmid vector construction, plant and animal subcellular localization, protein interaction analysis, to functional validation.
Strict Experimental Standards: We ensure technical reliability and data reproducibility throughout the entire experimental process, supporting publication-ready images compliant with SCI journal guidelines.
Application Scope
Determine the intracellular localization of biological macromolecules such as proteins, which helps to infer their potential involvement in biological processes and functions. This is applied in research fields including protein function studies, crop genetic improvement, drug target discovery, and disease mechanism exploration.
Sample Requirements
Type | Submission Criteria | Transport Conditions |
Sequence for Gene Synthesis | None | None |
Constructed Plasmid; Intermediate plasmid | Concentration > 100 ng/µl, volume > 20 µl; please provide the vector sequence and antibiotic resistance marker; must be free of contamination. | Ice pack |
Bacterial culture containing the constructed plasmid; Bacterial culture containing the intermediate plasmid | Volume > 500 µl; please provide the vector sequence and antibiotic resistance marker; must be free of contamination. | Dry ice shipment: 1 day (up to 5 kg), 2–3 days (10–20 kg), 3–5 days (20–30 kg). |
Plate Bacteria | No contamination | Room temperature |
Scope of Services and Delivery Standards
When using tobacco as the experimental material:
Category | Service Content | Working days | Deliverables |
Tobacco System Subcellular Localization | 1.Gene Synthesis / Vector Construction ☐Gene synthesis (codon optimized) Gene cloned into pBinGFP2 / pCAMBIA1300-mCherry-Flag vectors
| 10 | 1.One aliquot of recombinant plasmid 2. Sequencing results of the recombinant plasmid 3.Raw experimental images: For co-localization: 5 images each for the target gene and empty vector control (including: fluorescence of target protein, fluorescence of marker, merged fluorescence of target protein and marker, bright field, and full merge) For regular localization: 3 images each for the target gene and empty vector control (including: fluorescence of target protein, bright field, and merged image) 4.Standard experimental report (including experimental procedures and workflow) |
2.Tobacco Leaf Transformation System (1)Agrobacterium transformation and PCR-based positive clone verification (2) Plant cultivation and Agrobacterium-mediated transient transformation of tobacco plants | 17 | ||
3.Result Observation, Imaging, and Analysis (1) Confocal imaging and photography | 3 |
When using protoplasts as the experimental material:
Category | Service Content | Working days | Deliverables |
Protoplast-Based Subcellular Localization System | 1. Gene Synthesis / Vector Construction ☐ Gene synthesis (codon optimized) ☐Gene synthesis (non-codon optimized) ☐ Vector construction Gene cloned into pUC19-35S-EGFP vector | 10 | 1. One aliquot of recombinant plasmid 2. Sequencing results of the recombinant plasmid 3. Raw experimental images: For co-localization: 5 images each for the target gene and empty vector control (including: target protein fluorescence, marker fluorescence, merged fluorescence of target protein and marker, bright field, and full merged image) For standard localization: 3 images each for the target gene and empty vector control (including: target protein fluorescence, bright field, and merged image) 4.Standard experimental report (including experimental steps and workflow) |
2. Plasmid Extraction 3.(Optional) Protoplast Transformation System ☐ Rice Protoplast Transformation System ☐ Arabidopsis Protoplast Transformation System ☐ Tobacco Protoplast Transformation System | 17 | ||
4.Result Observation, Imaging, and Analysis (1) Confocal microscopy imaging (2) Data processing and analysis | 3 |
When using mammalian 293T cells as the experimental material:
Category | Service Content | Working days | Deliverables |
293T Cell Subcellular Localization | 1. Vector Construction Work A. Gene optimization and synthesis B. Subcloning into the expression vector | 15 | 1. Final report and all raw data (including experimental procedures, reagents, and equipment) 2. Return of remaining plasmid of the target gene 3. Delivery of images from 3 fields of view, each containing 2 fluorescence images (marker and target protein), 1 bright field image, and 1 merged image — a total of 4 images per field
|
2. Cell Transfection A. Cell passaging and plating B. Cell transfection C.Transient expression | 5 | ||
3. Result Observation, Imaging, and Analysis A. Cell fixation B.Fluorescence imaging C.Data organization and analysis | 4 |
Case Results Study
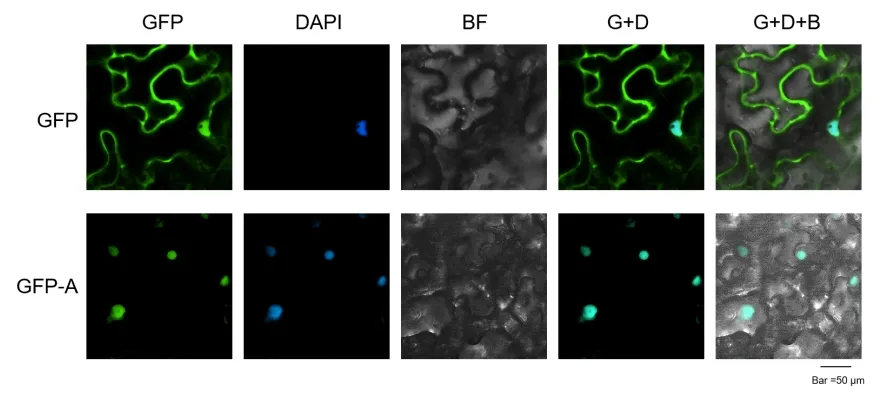
Example images of subcellular localization in the tobacco leaf system (Protein A localized in the nucleus)
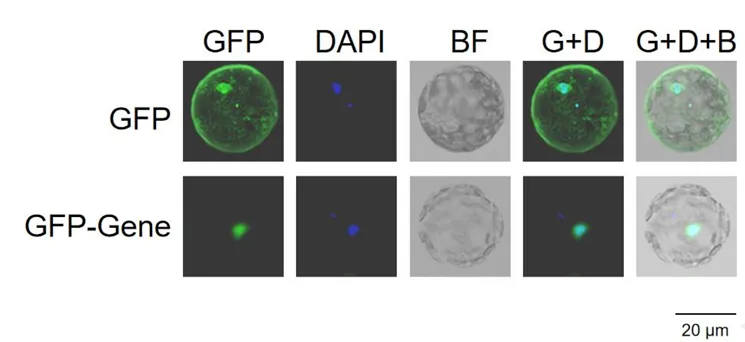
Example images of subcellular localization in the Arabidopsis protoplast system (Gene protein localized in the nucleus)
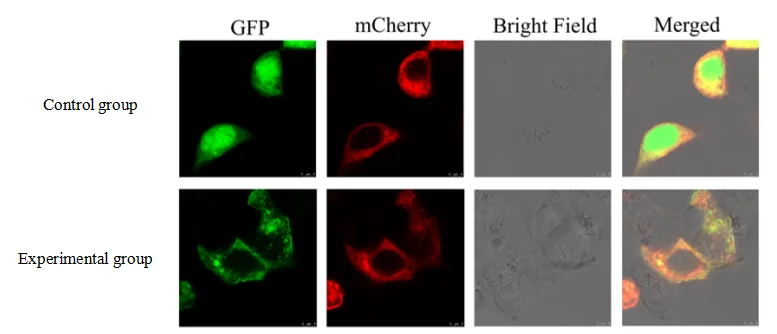
Example images of subcellular localization in mammalian 293T cells (target protein possibly localized in the cell membrane)
References
[1] Chaturvedi NK, Mir RA, Band V, et al. Experimental validation of predicted subcellular localizations of human proteins. BMC Res Notes. 2014 Dec 15;7:912.
[2] Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007 Sep;51(6):1126-36.
[3] Pagny S, Bouissonnie F, Sarkar M, et al. Structural requirements for Arabidopsis beta1,2-xylosyltransferase activity and targeting to the Golgi. Plant J. 2003 Jan;33(1):189-203.
[4] Chakrabarty R, Banerjee R, Chung SM, et al. PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant Microbe Interact. 2007 Jul;20(7):740-50.
Get Started with Our Subcellular Localization Services
Ready to pinpoint your protein's functional site within the cell? Contact us below to discuss your project or request a customized quote. Our team is here to support your research with accurate, publication-ready results.
Statement: Our services are exclusively for research applications and are not intended for diagnostic or therapeutic use.
Services Workflow

Online Consultation
01

Solution Matching
02

Service Contract
03

One-Stop-Services
04

Project Report
05
Related Products
Product Inquiry