Literature Sharing | Two pathogen-inducible UDP-glycosyltransferases, UGT73C3 and UGT73C4, catalyze the glycosylation of pinoresinol to promote plant immunity in Arabidopsis
Release time:
2025-07-01
This study uncovers a novel immune regulatory pathway in Arabidopsis thaliana involving two UDP-glycosyltransferases, UGT73C3 and UGT73C4, which are highly induced by Pseudomonas syringae infection. Overexpression of these genes enhances disease resistance, while their double mutation compromises immunity. Metabolomic analysis and biochemical assays reveal that UGT73C3/C4 glycosylate pinoresinol into its mono- and diglucoside forms, which promote immune responses by boosting ROS production and callose deposition. Additionally, the transcription factor HB34 directly activates UGT73C3/C4 expression, linking transcriptional regulation to glycosylation-mediated immunity. This work highlights the physiological significance of UGTs in plant defense through pinoresinol glycosylation.

This section reports that UGT73C3 and UGT73C4 were identified as pathogen-responsive genes through microarray data analysis, showing strong induction upon Pseudomonas syringae (Pst DC3000 and Pst DC3000-AvrRpm1) infection. Phylogenetic analysis confirmed their close relationship within the UGT73C subfamily, all members sharing the conserved PSPG box typical of UGTs. RT–qPCR and GUS reporter assays further validated their pathogen-inducible expression. These findings suggest that UGT73C3 and UGT73C4 are likely involved in plant immune responses.
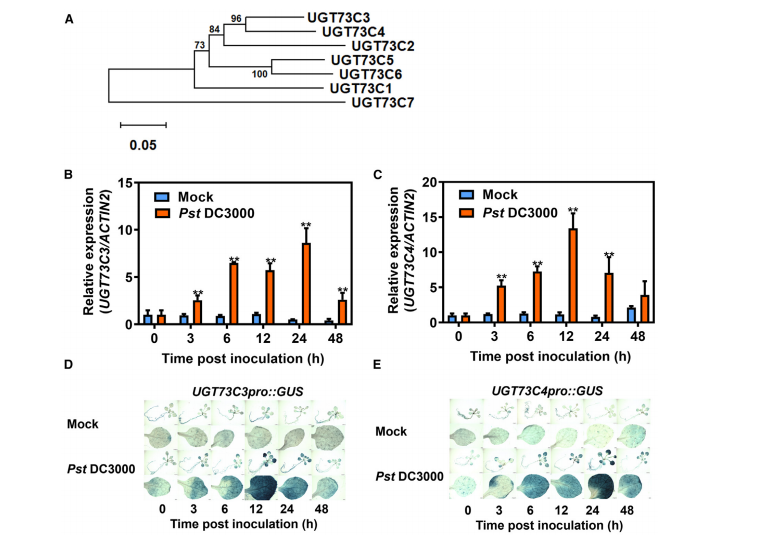
Figure 1. Phylogenetic analysis and induction of UGT73C3 and UGT73C4 expression by Pst DC3000.
To investigate the immune function of UGT73C3 and UGT73C4, researchers generated overexpression (OE) lines and CRISPR/Cas9 knockout mutants in Arabidopsis. Overexpression lines (UGT73C3OE and UGT73C4OE) exhibited enhanced resistance to Pseudomonas syringae (Pst DC3000), showing reduced bacterial growth and no necrotic lesions compared to wild type (WT). In contrast, single mutants (ugt73c3 or ugt73c4) showed no significant change in pathogen susceptibility. However, double mutants (ugt73c3ugt73c4) displayed more severe disease symptoms and higher bacterial loads, indicating functional redundancy between the two genes. Consistently, compensatory upregulation of UGT73C3 in ugt73c4 mutants and UGT73C4 in ugt73c3 mutants further supports their overlapping roles in promoting plant immunity.
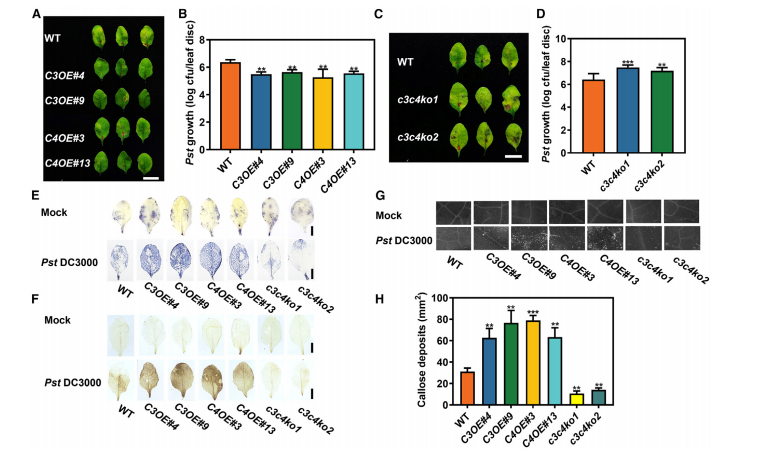
Figure 2. UGT73C3 and UGT73C4 enhance plant resistance to pathogens
To identify the catalytic substrates and metabolic functions of UGT73C3 and UGT73C4, researchers performed comprehensive secondary metabolome profiling on pooled overexpression (C3/C4OE) and wild-type (WT) Arabidopsis samples after Pst DC3000 infection. UPLC–MS/MS analysis detected 1202 metabolites across diverse chemical classes. Principal component analysis (PCA) effectively separated C3/C4OE from WT, indicating distinct metabolic profiles. A total of 36 differential metabolites (17 upregulated, 19 downregulated) were identified in C3/C4OE lines. Notably, pinoresinol diglucoside (PDG) was the most significantly upregulated glycoside, along with increases in several other glycosylated compounds. These findings suggest that UGT73C3/C4 likely catalyze pinoresinol glycosylation and contribute to changes in secondary metabolite profiles, potentially linking their enzyme activity to enhanced plant immunity.
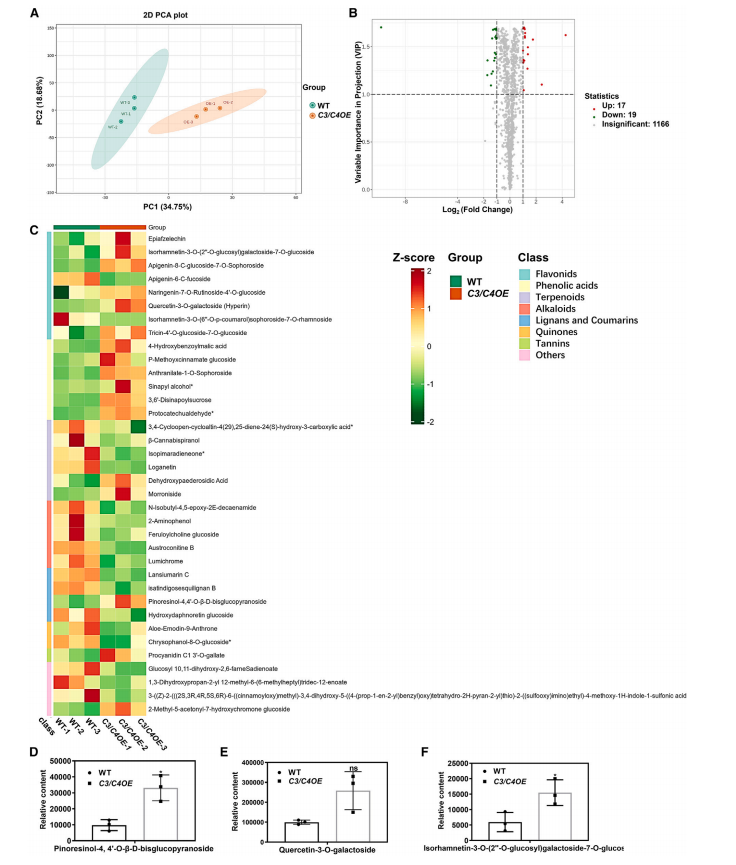
Figure 3. Analysis of secondary metabolites in WT and overexpression lines after Pst DC3000 inoculation.
This study confirms the catalytic activity of UGT73C3 and UGT73C4 toward pinoresinol. Using recombinant GST fusion proteins, the enzymes converted pinoresinol into two glycosylated products, pinoresinol monoglucoside (1a) and pinoresinol diglucoside (1b), with UDP-glucose as the sugar donor. LC–MS and LC–MS/MS analyses validated these products by their mass-to-charge ratios and fragmentation patterns, confirming glycosylation. Compared to other substrates such as quercetin, apigenin, and naringenin, UGT73C3/C4 showed higher activity and affinity for pinoresinol, indicating it as their preferred substrate. Furthermore, pinoresinol strongly induced the expression of UGT73C3 and UGT73C4, supporting their functional link to pinoresinol metabolism.
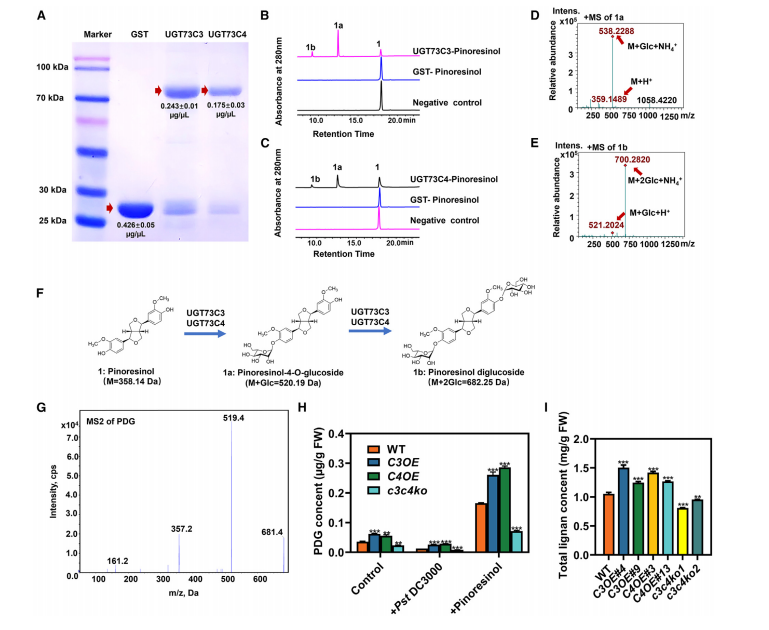
Figure 4. In vitro enzyme activities of UGT73C3 and UGT73C4 and in vivo measurements of glycoside.
To confirm the role of pinoresinol diglucoside (PDG) in plant defense, PDG pretreatment was applied at different concentrations before infection with Pst DC3000. Higher PDG levels correlated with reduced disease symptoms and bacterial growth at 3 days post-inoculation. While PDG alone did not trigger reactive oxygen species (ROS) or callose deposition, the combination of PDG and Pst DC3000 significantly increased the accumulation of superoxide (O2−), hydrogen peroxide (H2O2), and callose compared to infection alone. This suggests that PDG enhances pathogen resistance by boosting ROS and callose responses during infection. Moreover, PDG pretreatment rescued the increased susceptibility of the ugt73c3ugt73c4 double mutants, indicating that PDG deficiency underlies their pathogen sensitivity.
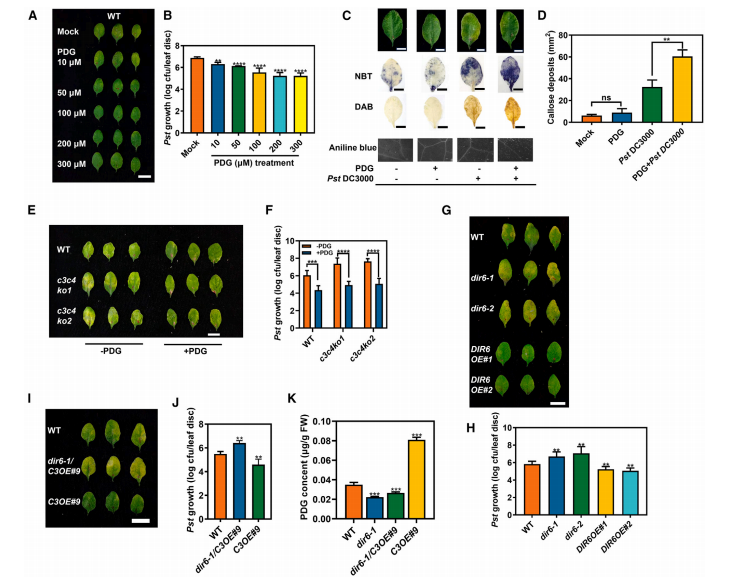
Figure 5. Pinoresinol diglucoside enhances plant resistance to Pst DC3000 infection.
To explore the transcriptional regulation of UGT73C3 and UGT73C4, a yeast one-hybrid screen identified HB34, a zinc finger homeodomain transcription factor, as a strong binder to their promoters. Subcellular localization studies showed that HB34-GFP is localized exclusively in the nucleus, supporting its role as a transcription factor. Bioinformatic analysis predicted the TAATTA motif as HB34’s binding site, and multiple such motifs were found in the promoters of UGT73C3 and UGT73C4, indicating that HB34 directly activates their expression.
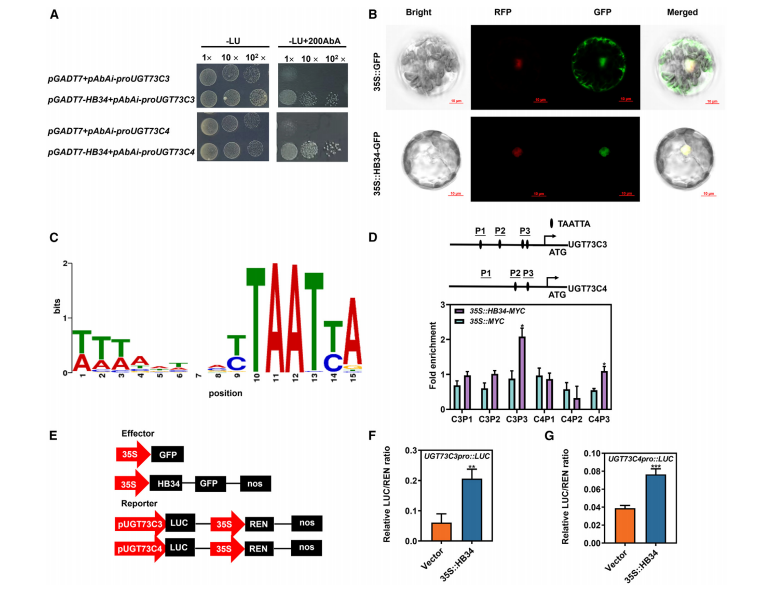
Figure 6. HB34 positively regulates UGT73C3 and UGT73C4 transcription by physically interacting with their promoters.
Chromatin immunoprecipitation followed by RT-qPCR showed that HB34 binds specifically to the P3 promoter fragments of UGT73C3 and UGT73C4 in transgenic plants overexpressing HB34-MYC, suggesting HB34’s involvement in their transcriptional regulation. Dual-luciferase assays demonstrated that HB34 overexpression significantly activated transcription from the UGT73C3 and UGT73C4 promoters. Further, expression analysis in HB34 overexpression and mutant lines confirmed that HB34 positively regulates UGT73C3 and UGT73C4 expression, establishing HB34 as an upstream transcriptional activator of these genes.
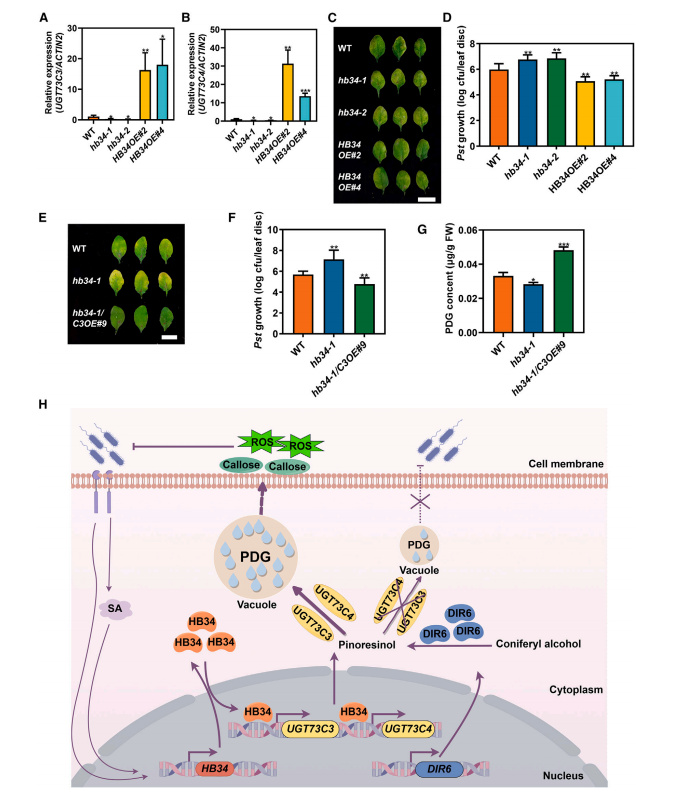
Figure 7. HB34 enhances plant resistance to pathogens.
Related News
2025-07-01
2025-06-27
2025-06-24
2025-06-20
2025-06-17
2025-06-12
2025-06-10
2025-06-06
Literature Sharing | SPL1 positively regulates cuticular ridge wax biosynthesis in Arabidopsis



