Literature Sharing | Genome-wide analysis of TGACG motif-binding factor (TGA) family and mechanism of PpTGA2.1-L and PpTGA7-L response to salicylic acid regulating fruit senescence through interaction with PpNPR1 in sand pear
Release time:
2025-06-10
This study investigates the roles of two TGA transcription factors, PpTGA2.1-L and PpTGA7-L, in the regulation of sand pear fruit senescence through the salicylic acid (SA) signaling pathway. Fourteen TGA genes were identified in sand pear, and phylogenetic analysis grouped them into five subfamilies. Transcriptomic data revealed that PpTGA2.1-L and PpTGA7-L were preferentially expressed during fruit senescence. SA treatment upregulated PpTGA2.1-L and downregulated PpTGA7-L. Yeast two-hybrid and luciferase assays confirmed interactions between both proteins and the SA receptor PpNPR1. Functional analyses showed that PpTGA2.1-L, in coordination with PpNPR1, delays fruit senescence by enhancing antioxidant enzyme activities, increasing SA levels, and reducing the accumulation of senescence-related markers. In contrast, PpTGA7-L promotes senescence by antagonizing PpNPR1. SA treatment mitigates the senescence effects mediated by PpTGA7-L. Overall, the study reveals that PpTGA2.1-L and PpTGA7-L have opposing roles in regulating pear fruit senescence via interactions with NPR1 in the SA signaling pathway.
In this study, a total of 14 TGA transcription factors were identified in the sand pear genome through HMM and BLAST analyses. Two of these, PpTGA2.1-L and PpTGA7-L, were previously identified from transcriptome sequencing of the cultivar 'Whangkeumbae' , and showed high sequence similarity (99.78% and 98.67%, respectively) with corresponding genomic sequences. Comparative analysis with Arabidopsis TGA proteins, which are grouped into five subfamilies, revealed that the 14 sand pear TGAs also fall into five groups. Specifically, PpTGA2.1-L belongs to Group II, and PpTGA7-L belongs to Group III.
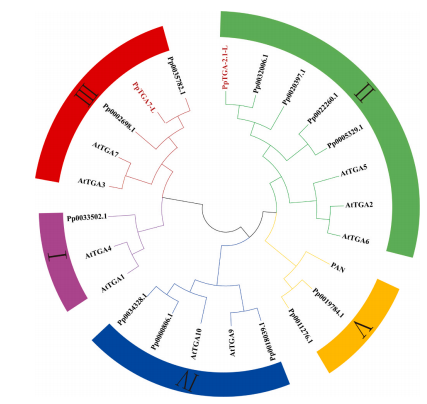
Fig. 1. Phylogenetic tree of TGA protein family based on Arabidopsis and sand pear genomes.
To explore the potential roles of PpTGA genes in hormone signaling, cis-element analysis was performed on their promoter regions. Seven PpTGA genes, including PpTGA2.1-L and PpTGA7-L, contained salicylic acid (SA)-responsive elements, suggesting their involvement in the SA pathway. Most genes also had abscisic acid-responsive elements, except for four. Five genes harbored auxin-responsive elements, while all except three contained jasmonic acid-responsive elements. Additionally, all PpTGA genes except two had gibberellin-responsive elements. These results indicate that PpTGA genes may participate in multiple hormone signaling pathways.
Expression analysis in the sand pear cultivars “Whangkeumbae” and “Akizuki” showed that PpTGA2.1-L and PpTGA7-L are widely expressed. PpTGA2.1-L is most highly expressed in shoots, while PpTGA7-L shows highest expression in petals and moderate expression in fruit flesh. During fruit maturation and postharvest stages, both genes peaked during senescence—PpTGA2.1-L at 25 days after harvest (DAH) and PpTGA7-L at 10 DAH.
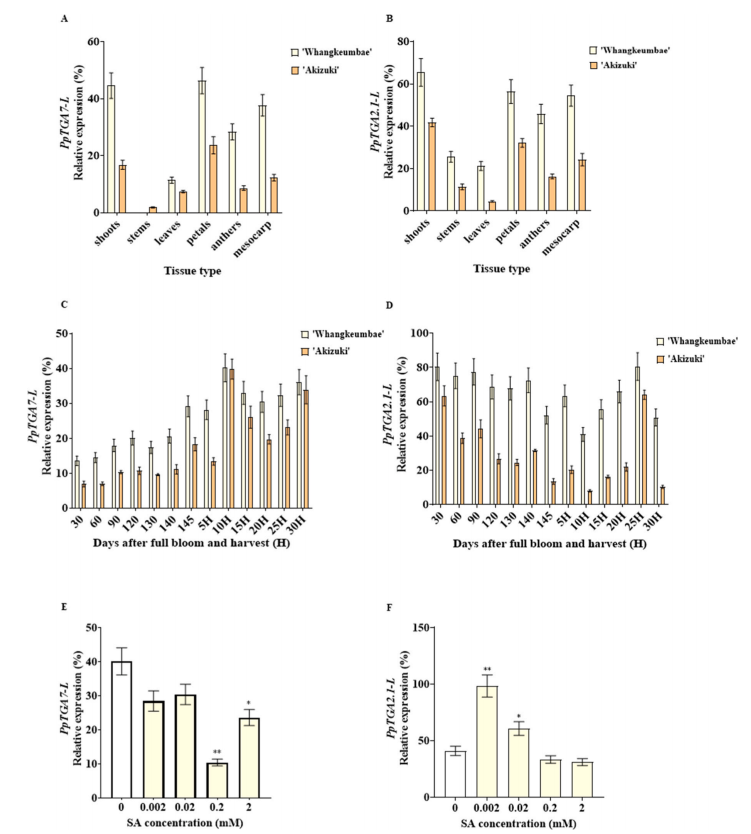
Fig. 2. The expression patterns of PpTGA2.1-L and PpTGA7-L in “Whangkeumbae” and “Akizuki”
TGA transcription factors are well-known for their roles in plant defense, but their function in fruit senescence remains unclear. Previous transcriptome analysis identified two salicylic acid (SA)-induced genes, PpTGA2.1-L and PpTGA7-L. Protein interaction predictions suggested both may interact with the SA receptor protein PpNPR1. These interactions were confirmed through yeast two-hybrid (Y2H) and luciferase complementation (LUC) assays, demonstrating that both PpTGA2.1-L and PpTGA7-L physically interact with PpNPR1.
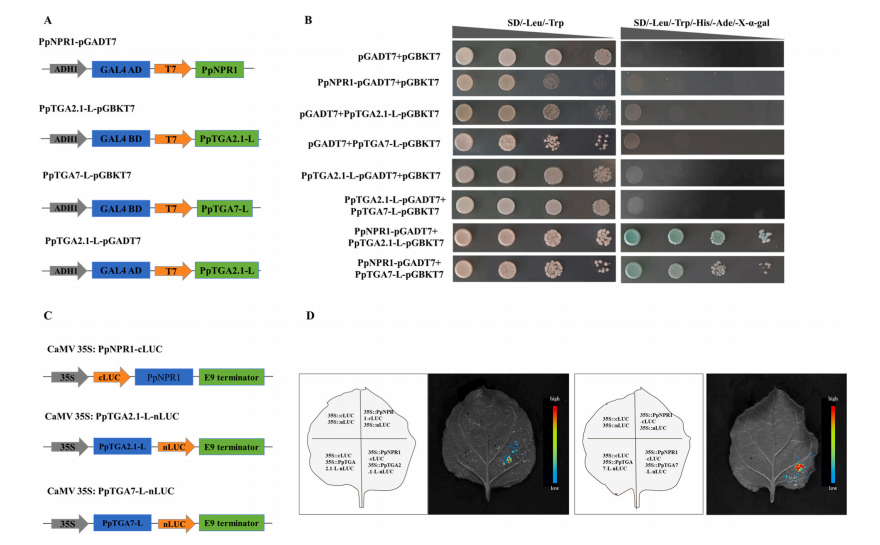
Fig. 3. PpTGAs interact with PpNPR1. PpTGA2.1-l and PpTGA7-L were connected to the pGBKT7 vector to form decoy protein by seamless cloning method.
To investigate the regulatory role of PpNPR1 in fruit senescence, its expression was analyzed across tissues and developmental stages. PpNPR1 showed highest expression in shoots, followed by fruit flesh. During fruit development, its expression peaked at 60 days after full bloom (DAFB), and during senescence, it was highest at 5 days after harvest (DAH).
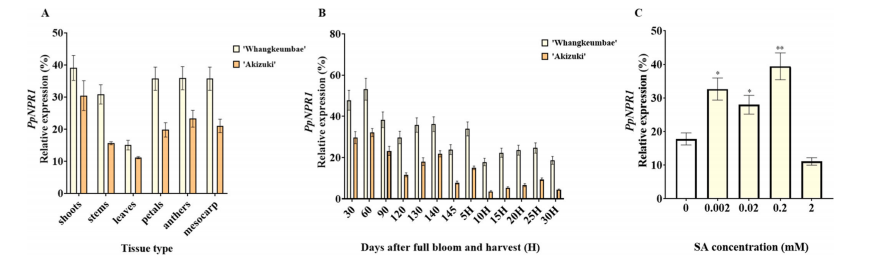
Fig. 4. Expression levels of PpNPR1 in shoots, stems, leaves, petals, anthers, and mesocarp of ‘Whangkeumbae’ and ‘Akizuki’
To study the roles of PpTGA2.1-L, PpTGA7-L, and PpNPR1 in pear fruit senescence and their response to salicylic acid (SA), these genes were silenced in mature ‘Akizuki’ fruit, followed by SA treatment. qRT-PCR confirmed effective gene silencing. After SA treatment, the expression of PpTGA2.1-L and PpNPR1 was upregulated, while PpTGA7-L expression was suppressed.

Fig. 5. Agrobacterium-mediated transient transformation was employed to silence PpTGA2.1-L, PpTGA7-L, and PpNPR1 in ‘Akizuki’ fruit
Silencing PpTGA2.1-L and PpNPR1 individually led to faster pear fruit senescence, shown by reduced firmness, increased ethylene production, lowered antioxidant enzyme activities (POD, SOD, CAT), higher cell wall-degrading enzyme activities (PG, PME, CEL), and decreased SA content. SA treatment delayed this senescence. Co-silencing both genes further accelerated senescence. Gene expression analysis revealed that silencing reduced antioxidant genes (PpPOD1, PpSOD1, PpCAT1) and increased senescence-related genes (PpPG1, PpPME2, PpCEL3), while SA treatment reversed these effects.
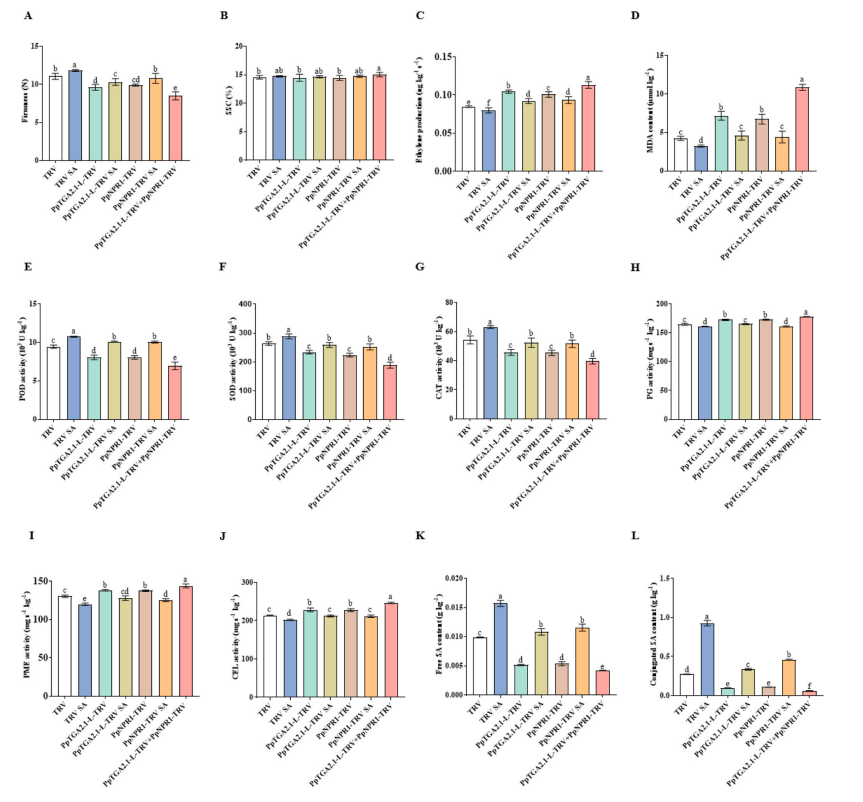
Fig. 6. The effects of individually silencing PpTGA2.1-L and PpNPR1
This study identified 14 TGA genes in sand pear, focusing on PpTGA2.1-L and PpTGA7-L, which show different expression patterns and opposite responses to salicylic acid (SA). Both interact with the SA receptor PpNPR1 but have contrasting roles: PpTGA2.1-L with PpNPR1 delays fruit senescence, enhanced by SA treatment, while PpTGA7-L antagonizes this effect. These results reveal how TGA genes and NPR1 co-regulate fruit senescence through the SA signaling pathway, offering valuable insights for future research and practical strategies to extend fruit shelf life.
Related News
2025-06-12
2025-06-10
2025-06-06
Literature Sharing | SPL1 positively regulates cuticular ridge wax biosynthesis in Arabidopsis
2025-06-04
Functional study of plant hormone transporters using a heterologous system
2025-05-29
2025-05-27
2025-05-22
2025-05-20



