Literature Sharing | SPL1 positively regulates cuticular ridge wax biosynthesis in Arabidopsis
Release time:
2025-06-06
This study investigates the molecular mechanisms underlying the formation and maintenance of cuticular ridge structures, which are characteristic of the sepals and petals in various plant species. The authors reveal that the production of cuticular ridge wax in Arabidopsis is regulated by SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors. Double mutants of SPL1 and SPL12 (spl1spl12) show severe defects in cuticular ridge formation, with significantly reduced seed-setting rates and increased organ fusion compared to wild-type plants. Further analysis demonstrates that SPL1 positively regulates the expression of key enzymes involved in wax biosynthesis, including the alkane-forming enzymes ECERIFERUM1 (CER1), CER1-like (CER1L), and the primary (1-) alcohol-forming enzyme CER4. These enzymes are essential for the synthesis of alkanes and 1-alcohols, which are crucial for the accumulation of cuticular ridge wax. Additionally, the study identifies HSP70–16 as an interacting partner of SPL1, enhancing its transcriptional activity. Together, these results uncover a regulatory network critical for cuticular ridge formation and provide new insights into plant developmental processes.

In the spl1spl12 double mutants, vegetative growth appeared normal, but there was a significant reduction in seed set during the post-reproductive phase due to flowers failing to produce siliques. Stereomicroscopy of stage 13 flowers revealed that the sepals of spl1spl12 mutants did not open properly compared to wild-type plants. Internal analysis of floral organs showed that stamens, gynoecia, and petals in the fused flowers did not extend properly, often growing twisted or curved. The restricted space in unopened buds prevented mature pollen from contacting the stigma. Additionally, papilla cells on the stigma were abnormal, leading to reduced pollination. However, pollen grains were viable at anthesis, and artificial self-pollination restored normal seed production, indicating that the male and female gametophytes were not defective. These results suggest that the reduced fertility in spl1spl12 is mainly due to the failure of sepals to open.

Fig. 1. Loss of sepal cuticular ridges wax in spl1spl12 reduces seed setting rate.
To investigate the underlying mechanisms, the researchers compared the transcriptomes of Col-0 and spl1spl12 flowers under normal conditions. KEGG annotation of differentially expressed genes (DEGs) revealed significant enrichment in pathways related to cutin, suberine, and wax biosynthesis, suggesting that the loss of SPL1 and SPL12 disrupts wax biosynthesis, leading to reduced wax accumulation in the sepal epidermis. Among the downregulated DEGs, key wax biosynthesis genes—CER1-Like (CER1L), CER1, and CER4—were notably reduced in spl1spl12 mutants. These CER genes showed higher expression levels in floral organs than in vegetative tissues, peaking at flower development stage 12. CER1L and CER1 are involved in the alkane-forming pathway of floral wax biosynthesis, while CER4, an acyl-CoA reductase, is crucial for primary alcohol formation. Mutations in CER4 lead to a significant reduction in primary alcohols and wax esters, further implicating it in the wax biosynthesis process.
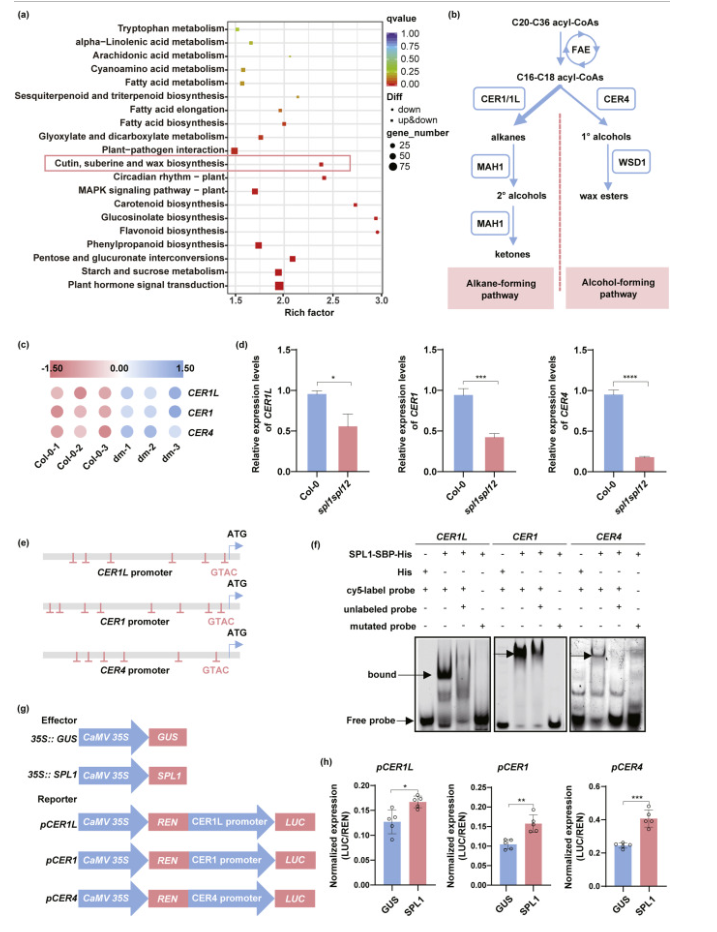
Fig. 2. SPL1 activates wax biosynthetic genes.
To identify potential interacting partners of SPL1, a yeast two-hybrid screening was performed, which identified 21 candidate proteins. Notably, three independent clones encoded AT1G11660, corresponding to the molecular chaperone HSP70–16. This is significant because previous studies have shown that loss-of-function mutants of HSP70–16 exhibit impaired reproductive development, including reduced seed set due to postgenital sepal fusion defects. Subcellular localization analysis revealed that SPL1 and HSP70–16 co-localize in N. benthamiana leaf epidermal cells. The physical interaction between SPL1 and HSP70–16 was confirmed using yeast two-hybrid assays, luciferase complementation, and bimolecular fluorescence complementation. A triple mutant (spl1spl12hsp70–16) was created, and phenotypic analysis showed that it exhibited more severe developmental defects than the spl1spl12 double mutant, including reduced seed set and abnormal cuticular ridge formation at the sepal tips. These results suggest a functional synergy between SPL1 and HSP70–16 in regulating reproductive development.
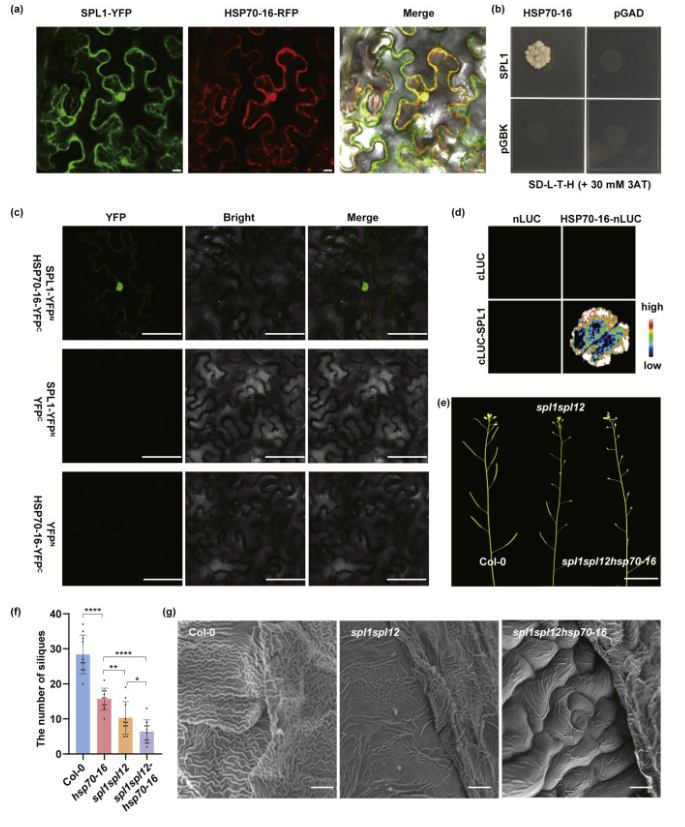
Fig. 3. SPL1 interacts with HSP70–16.
Comparative transcriptomic profiling of the spl1spl12hsp70–16 triple mutants versus wild-type plants, followed by KEGG pathway enrichment analysis, revealed significant differential expression of genes linked to lipid metabolism, including fatty acid biosynthesis, elongation, and pathways related to cuticular wax and suberin biosynthesis. This transcriptional reprogramming suggests a synergistic regulatory interaction between SPL1 and HSP70–16 in modulating these metabolic processes.
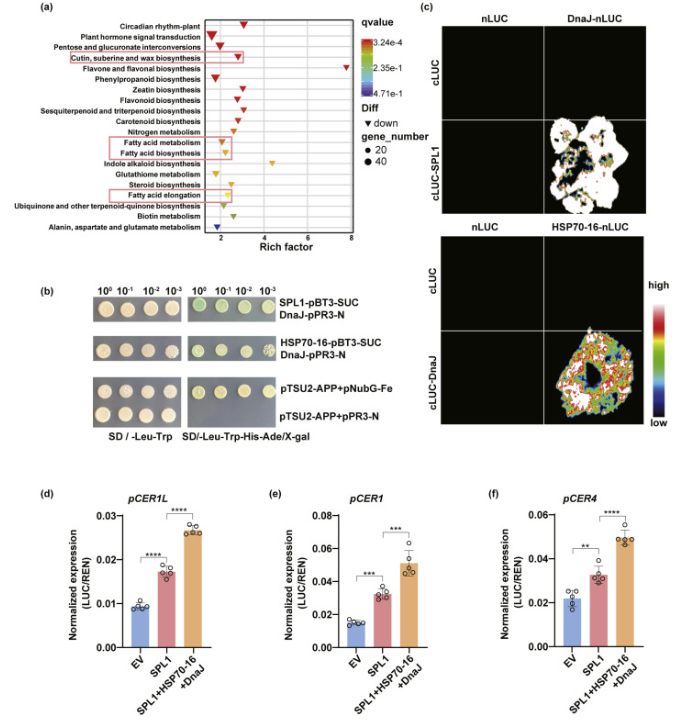
Fig. 4. HSP70–16 enhances SPL1 transcriptional activity through protein interaction.
In summary, the study reveals the critical role of SPL proteins in cuticular ridge formation. The spl1spl12 double mutants show reduced wax in sepal cuticular ridges, leading to sepal fusion, and failure in flowering and seed setting. These findings highlight the importance of an intact cuticle for plant growth and development. Further analysis shows that HSP70–16 interacts with SPL1, enhancing its transcriptional activation of wax biosynthesis genes. The authors propose a novel regulatory mechanism involving the SPL1-HSP70–16 module, which is crucial for sepal cuticular ridge formation and preventing organ fusion. This work provides insights into cuticle integrity and broader developmental processes in plants.
Related News
2025-06-06
Literature Sharing | SPL1 positively regulates cuticular ridge wax biosynthesis in Arabidopsis
2025-06-04
Functional study of plant hormone transporters using a heterologous system
2025-05-29
2025-05-27
2025-05-22
2025-05-20
2025-05-16
2025-05-13



