Literature Sharing | ERF100 regulated by ERF28 and NOR controls pectate lyase 7, modulating fig (Ficus carica L.) fruit softening
Release time:
2025-05-13
This study investigates the regulatory mechanisms of fig fruit textural changes during ripening. It focuses on the transcription factor FcERF100, which plays a key role in repressing fruit softening. FcERF100 transcription is rapidly suppressed during fig fruit ripening, and its overexpression delays softening by decreasing the expression of the cell wall-modifying gene FcPL7. Through various assays (Y1H, ChIP-qPCR, EMSA, and dual-luciferase), it was shown that FcERF100 represses FcPL7 by binding directly to its promoter via specific elements (GCC-box and DRE/CRT).
Additionally, FcERF28 was identified as an upstream regulator of FcERF100, activating it through direct binding. The study also uncovered the involvement of a NAC transcription factor, FcNOR, which interacts with FcERF100, forming a complex that reduces FcERF100’s repression of FcPL7. Silencing FcNOR retarded fruit softening and decreased FcPL7 expression. Furthermore, ethylene treatment was found to modulate the expression of these factors, enhancing FcNOR and FcPL7 levels while reducing FcERF28 and FcERF100. This research identifies a novel FcERF100-centered regulatory complex that controls the cell wall modification during early fruit growth and ripening.

Fig fruit undergoes visible size and color changes during development. In early stages (S1 and S2), the fruit is firm with low ethylene release and PL activity. At stage S3, as ripening begins, fruit firmness decreases by about 50%, ethylene release increases significantly, and PL activity starts to rise. Ethylene release peaks at S4, while PL activity continues to increase, reaching its highest at S5.
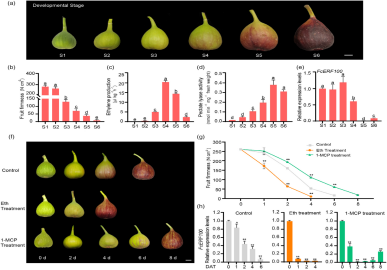
Figure 1 Ethylene promotes fig fruit softening.
Ethylene's effect on fig fruit firmness was tested using ethephon and 1-MCP treatments. Ethephon accelerated ripening and significantly decreased fruit firmness, while 1-MCP delayed ripening, coloration, and softening. These results confirm that ethylene enhances fig fruit softening during ripening.
FcERF100 was identified in a previous RNA-seq study as a key regulator of fig fruit softening. It is localized to the nucleus and its expression significantly decreases at the onset of ripening, with ethylene treatment inhibiting its expression. Transient overexpression of FcERF100 in pre-ripening fruit increased fruit firmness and reduced pectate lyase (PL) activity, while silencing FcERF100 resulted in decreased firmness and higher PL activity, confirming its negative regulatory role in fig fruit softening. No significant changes were observed in polygalacturonase (PG) or pectin methylesterase (PME) activities.
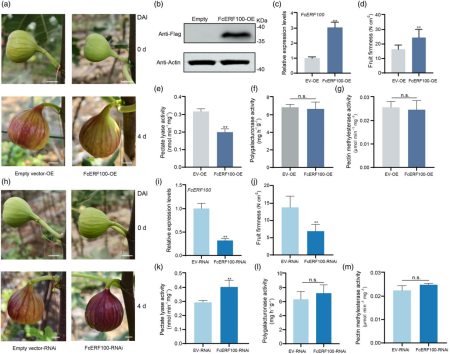
Figure 2 illustrates that FcERF100 represses fruit softening in fig. Panels (a) and (h) show fig fruit with transient overexpression of FcERF100 (FcERF100-OE), empty vector (EV-OE) as control, transient silencing of FcERF100 (FcERF100-RNAi), and empty vector (EV-RNAi) as control during the ripening process.
In this study, the FcERF100 gene was overexpressed in ‘Micro-Tom’ tomato to investigate its role in fruit softening. The transgenic lines (OE-1, OE-2, and OE-3) showed higher levels of FcERF100 expression compared to wild-type (WT) fruit. Four days after the breaker stage (BR+4), FcERF100-OE fruit exhibited higher firmness than the WT. Additionally, ethylene production, carotenoid content, and the activities of pectinase (PL) and polygalacturonase (PG) were significantly lower in FcERF100-OE fruit, while pectin methylesterase (PME) activity remained unchanged. Gene expression analysis revealed that the transcription of genes involved in lycopene biosynthesis (PSY1), fruit softening (PL, PG2a), and key ripening regulators (NOR, RIN) were downregulated in the FcERF100-OE lines compared to WT at both the BR and BR+4 stages.
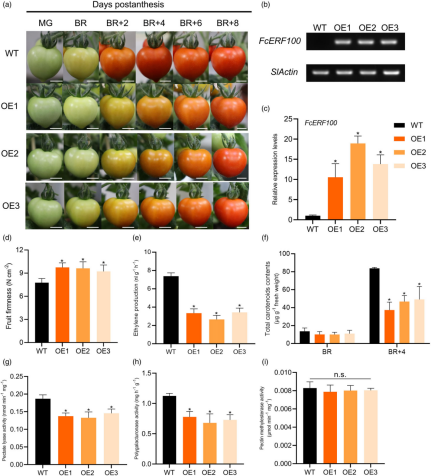
Figure 3 Overexpression of FcERF100 inhibits fruit softening in tomato. (a) Fruit of wild-type (WT) and FcERF100-overexpressing (OE) lines at late stages of development
This study explored the mechanism by which FcERF100 regulates the expression of FcPL7. The promoter analysis of FcPL7 revealed two GCC-box and one DRE/CRT cis-elements. Yeast one-hybrid (Y1H) assays demonstrated that FcERF100 directly binds to the FcPL7 promoter. Further validation using electrophoretic mobility shift assays (EMSA) showed that FcERF100 specifically binds to the FcPL7 promoter in vitro, requiring the GCC or DRE/CRT recognition sequences. Chromatin immunoprecipitation (ChIP)-qPCR confirmed this interaction in fig callus. Additionally, co-infiltration of FcERF100 and the proFcPL7–luciferase (LUC) reporter in Nicotiana benthamiana led to significantly reduced luminescence and LUC activity, indicating that FcERF100 negatively regulates FcPL7 expression.
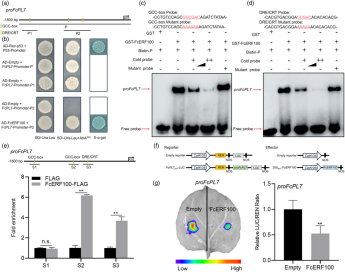
Figure 4 FcERF100 binds directly to FcPL7 promoter in vitro and in vivo
In this study, a Y1H library screening identified FcERF28 as a potential transcription factor (TF) regulating FcERF100 expression. Among 61 positive clones, FcERF28 (FCD\_00006831), which has been previously linked to fig fruit ripening and negatively regulated by ethylene, was identified. Further validation using the Y1H system confirmed that FcERF28 binds to the FcERF100 promoter. EMSA with purified His-labelled FcERF28 protein confirmed this binding in vitro. A dual-luciferase assay demonstrated that co-infiltration of FcERF28 and proFcERF100–LUC in Nicotiana benthamiana leaves resulted in higher LUC activity, indicating that FcERF28 positively regulates the promoter activity of FcERF100.
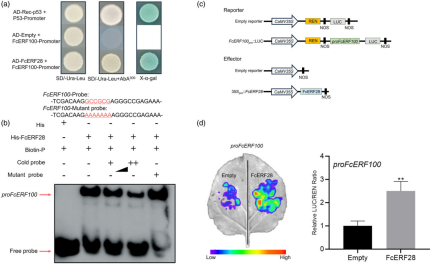
Figure 5 FcERF28 directly activates the transcription of FcERF100.
This study aimed to identify potential interacting partners of FcERF100 involved in its function. Using yeast two-hybrid (Y2H) screening with FcERF100 as bait, a NAC family protein was identified, which showed 54% amino acid homology to the global ripening regulator SlNOR. This protein was renamed FcNOR. Validation experiments confirmed that FcERF100 interacts directly with FcNOR. In vitro pull-down assays confirmed the interaction, as FcNOR was detected in the pulled-down samples with FcERF100 but not with His protein alone. LUC complementation imaging (LCI) assay and bimolecular fluorescence complementation (BiFC) assay both confirmed the interaction between FcERF100 and FcNOR in plant cells. These results collectively demonstrate that FcERF100 interacts with FcNOR.
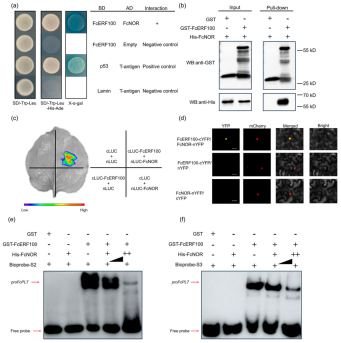
Figure 6 Physical interaction of FcNOR and FcERF100.
This study reveals a novel FcERF100-centered regulatory mechanism controlling fig fruit softening. In young fruit, low ethylene levels maintain high expression of FcERF28 and FcERF100, which together repress FcPL7 transcription, limiting pectin lyase (PL) activity. FcERF28 also enhances FcERF100 expression, forming a transcriptional cascade. As fruit ripens and ethylene levels rise, FcERF28 and FcERF100 expression declines, relieving repression on FcPL7. Concurrently, FcNOR expression increases and inhibits FcERF100 by forming a FcERF100–FcNOR complex, further lifting repression on FcPL7. This results in elevated PL activity and rapid fruit softening. These findings provide insight into the transcriptional regulation of fruit softening and suggest strategies for its precise control in climacteric fruits.
Related News
2025-05-13
2025-05-09
2025-05-07
2025-04-29
2025-04-25
2025-04-22
A Comprehensive Overview – Dual-Luciferase Reporter Gene Assay
2025-04-18
2025-04-15



