Literature Sharing | MADS-domain transcription factor AGAMOUS LIKE-9 participates in the gibberellin pathway to promote bud dormancy release of tree peony
Release time:
2025-05-07
This study explores the molecular mechanism underlying bud dormancy release in tree peony, with a particular focus on the gibberellin (GA) signaling pathway. Researchers identified PsAGL9, a MADS-box transcription factor, as an interacting partner of PsRGL1, a DELLA protein that negatively regulates dormancy release. PsAGL9 expression was induced by both chilling treatment and exogenous GA₃ application, indicating its potential role in the GA-mediated dormancy process. Protein–protein interaction assays, including yeast two-hybrid, pull-down, and luciferase complementation assays, confirmed that PsAGL9 physically interacts with PsRGL1. Overexpression of PsAGL9 significantly promoted bud dormancy release and upregulated key dormancy-related marker genes such as PsBG6, PsBG9, PsEBB1, PsEBB3, and PsCYCD. Further analyses revealed that PsAGL9 directly binds to the promoter regions of PsCYCD and PsEBB3 through classical and nonclassical CArG-box motifs, as demonstrated by yeast one-hybrid (Y1H), electrophoretic mobility shift assays (EMSA), and dual-luciferase reporter assays. However, PsRGL1 was found to inhibit the DNA-binding ability of PsAGL9, thus acting as a repressor of PsAGL9’s transcriptional activity. In addition to forming homodimers, PsAGL9 also interacts with other MADS-box proteins such as PsAGL6 and PsPI to form heterodimers, which further enhance the transcription of target genes. Altogether, this work reveals a novel regulatory module, PsRGL1–PsAGL9–PsCYCD, that functions in GA-mediated bud dormancy release, offering new insights into the hormonal and transcriptional regulation of dormancy in perennial woody plants.

In this study, PsRGL1, a DELLA protein lacking DNA-binding ability, was identified as a key negative regulator of bud dormancy release in tree peony. To uncover potential PsRGL1-interacting transcription factors, a GST-PsRGL1 fusion protein was used in pull-down assays with total proteins extracted from buds subjected to different chilling treatments. LC–MS/MS analysis revealed several candidate interactors, including two MADS-box proteins. BLAST analysis showed that these MADS-box proteins belonged to the SOC1 and AGL9 clades. While SOC1 is already known to promote endodormancy release, the other gene, named PsAGL9, encoded a 243-amino acid protein containing MADS- and K-box domains. Phylogenetic analysis confirmed its close relationship to PpAGL9 and MdAGL9. Expression analysis by qRT-PCR showed that PsAGL9 was induced by early chilling, continued to increase with prolonged chilling, and was also upregulated by exogenous GA₃, suggesting its role in the GA signaling pathway related to dormancy release. In situ hybridization revealed that PsAGL9 was highly expressed in stamens and petals after chilling, with transcript levels peaking at 7–14 days and then declining. Its expression was also highest in floral organs and lowest in roots during early flowering. These results indicate that PsAGL9 may play dual roles in regulating both floral organ development and bud dormancy release in tree peony.
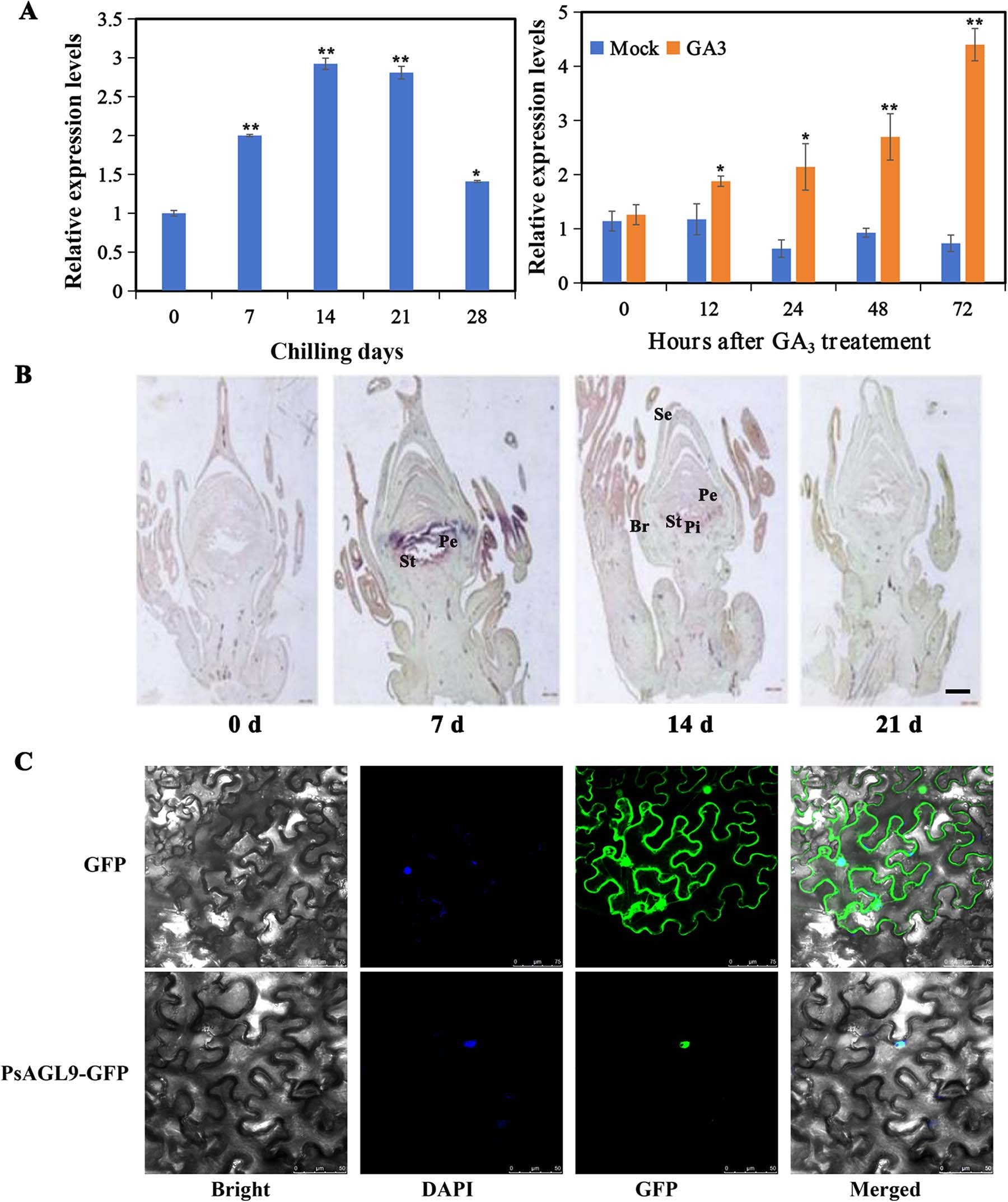
Figure 1. Expression of PsAGL9 and subcellular localization.
This paragraph confirms the physical interaction between PsAGL9 and PsRGL1 through multiple assays. A modified yeast two-hybrid (Y2H) assay, which used a truncated PsRGL1 to avoid self-activation, showed that PsAGL9 and PsRGL1 interact in yeast. This interaction was further validated in planta using a luciferase complementation assay in Nicotiana benthamiana, where co-expression of PsAGL9 and PsRGL1 produced strong fluorescence. Additionally, a pull-down assay using purified fusion proteins (GST-PsRGL1 and MBP-PsAGL9) also demonstrated a direct interaction. These results collectively confirm that PsAGL9 interacts with PsRGL1 and likely functions as part of the GA signaling pathway in regulating bud dormancy release.
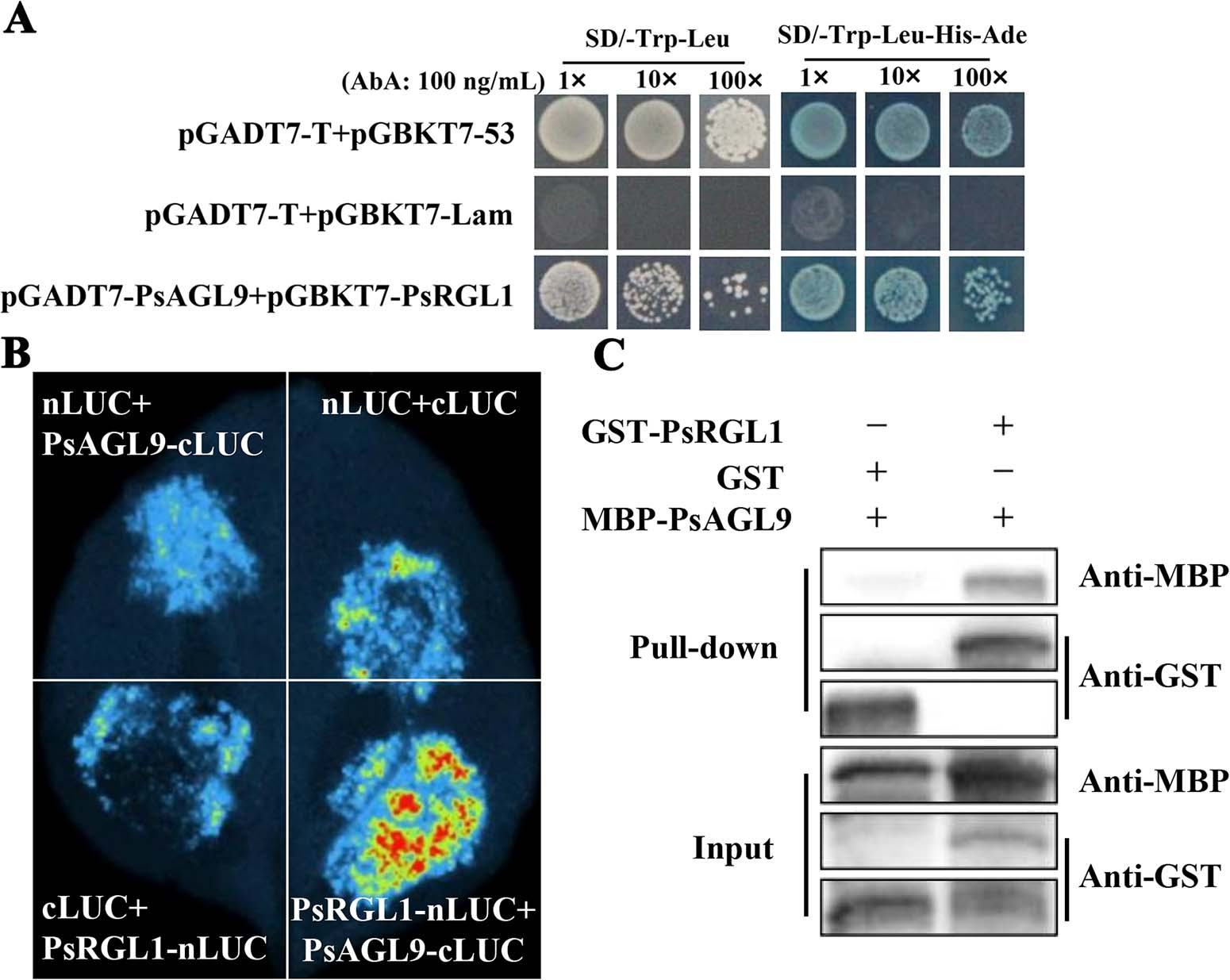
Figure 2. Identification of the interaction between PsAGL9 and PsRGL1.
To investigate the role of PsAGL9 in bud dormancy release, chilled tree peony buds were transformed with a pBI121-PsAGL9 overexpression construct. qRT-PCR analysis at 7 days after infection (DAI) confirmed significantly higher PsAGL9 expression in transformed buds compared to controls. In selected overexpression lines (PsAGL9-OE-7, 10, and 11), key dormancy-related genes (PsCYCD, PsEBB1, PsEBB3, PsBG6, and PsBG9) were strongly upregulated. Phenotypically, PsAGL9-overexpressing buds began breaking dormancy earlier and achieved an 85% budbreak rate by 15 DAI, while control buds showed delayed budbreak starting only at 20 DAI. These findings demonstrate that PsAGL9 promotes bud dormancy release in tree peony.
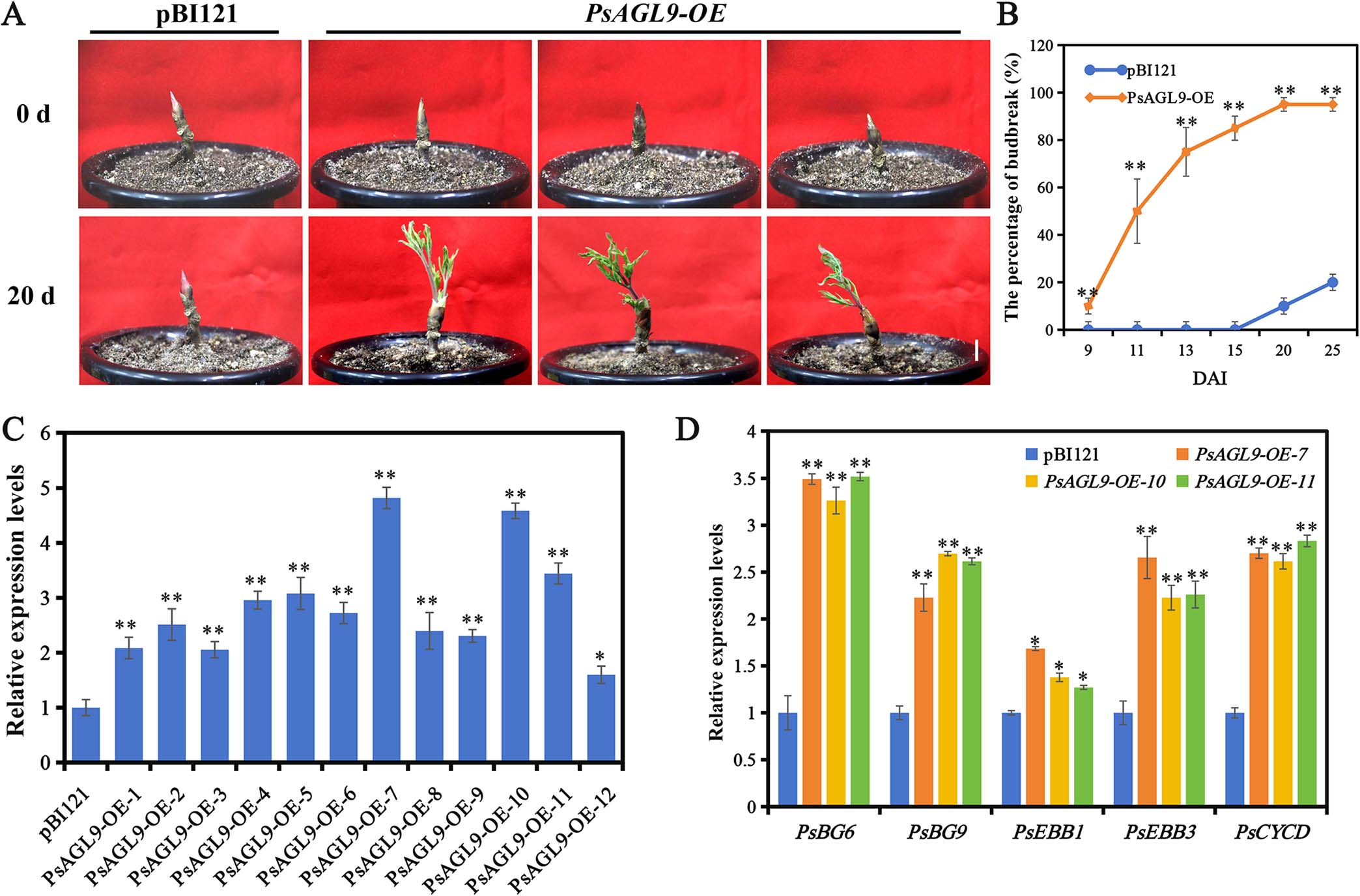
Figure 3. Overexpression of PsAGL9-promoted bud dormancy release in tree peony
Yeast one-hybrid (Y1H) assays demonstrated that PsAGL9 binds to the promoter of PsCYCD, specifically to the region containing a classical CArG motif. This interaction was further confirmed by EMSA, where the GST-PsAGL9 protein strongly bound a biotin-labeled DNA probe with the CArG motif, and the binding was competitively reduced by excess unlabeled probe but not by a mutant version. Additionally, dual-luciferase assays in tobacco leaves showed that coexpression of PsAGL9 significantly enhanced the activity of the PsCYCD promoter, confirming that PsAGL9 directly binds to and activates PsCYCD expression.
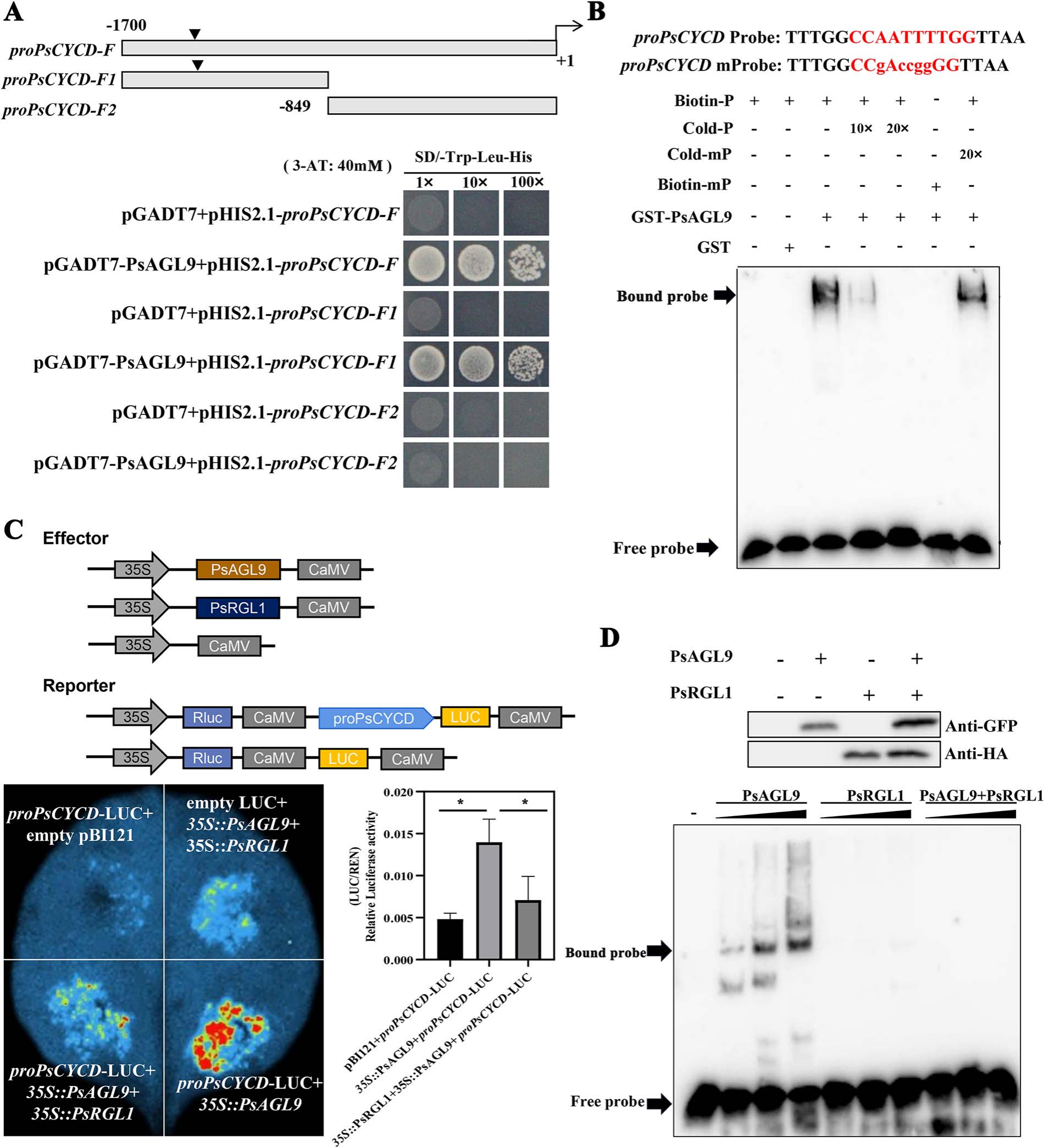
Figure 4 PsAGL9 directly activated the expression of PsCYCD by binding to its promoter.
A nonclassical CArG motif was identified in the PsEBB3 promoter, suggesting potential regulation by PsAGL9. Yeast one-hybrid (Y1H) assays confirmed that PsAGL9 bound to the promoter region containing this motif, and EMSA further validated the specific binding. Dual-luciferase assays demonstrated that PsAGL9 could activate PsEBB3 expression, indicating its direct regulatory role.
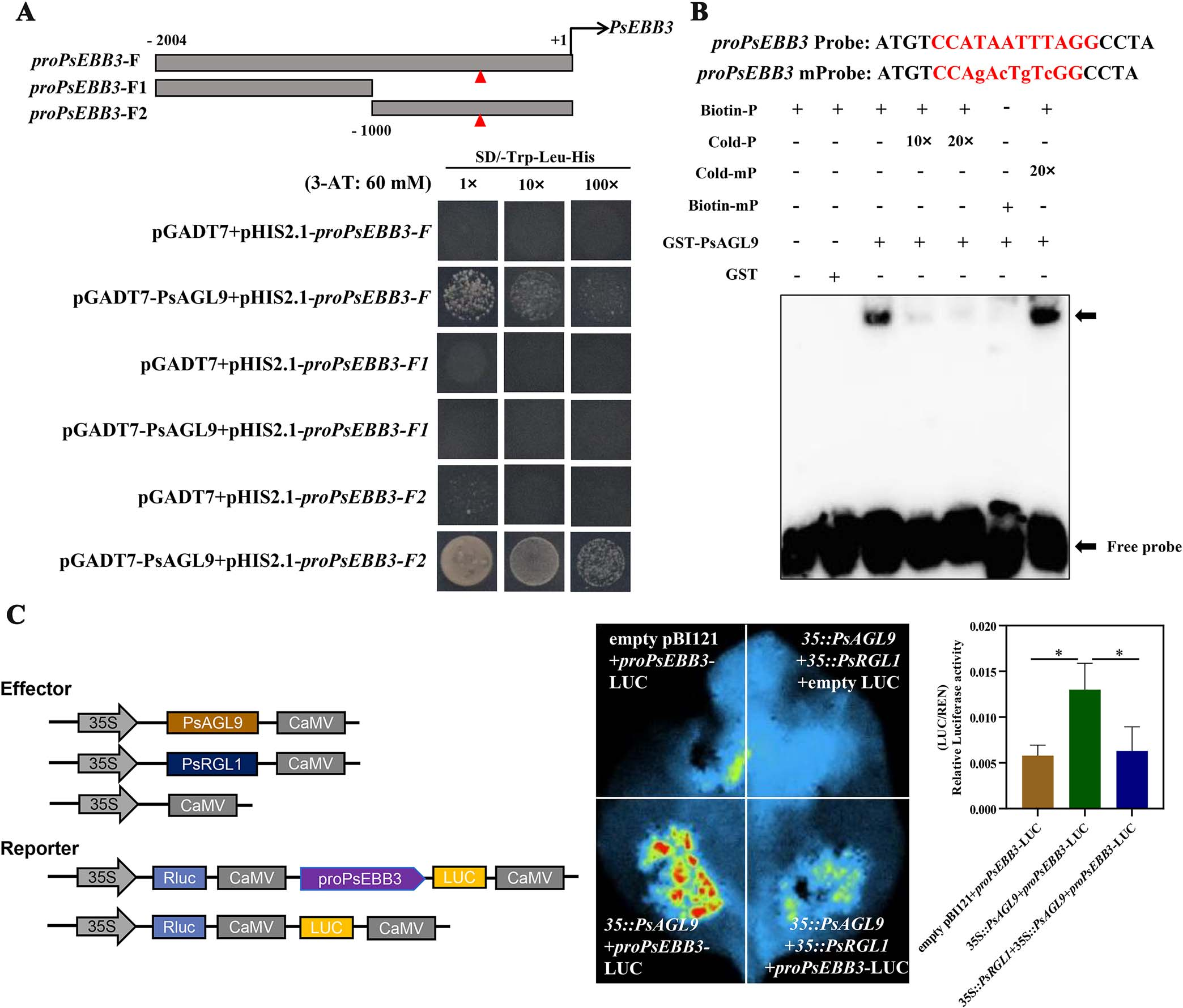
Figure 5. PsAGL9 directly bound and activated the expression of PsEBB3.
This study proposes a model in which PsAGL9 promotes bud dormancy release in tree peony. Initially, PsAGL9 is induced by chilling and GA₃ but is sequestered by the DELLA protein PsRGL1, preventing it from activating target genes. As chilling continues, endogenous bioactive GAs trigger PsRGL1 degradation, freeing PsAGL9 to form heterodimers with PsAGL6 or PsPI. These complexes bind to the promoters of PsCYCD and PsEBB3, activating their expression and promoting cell proliferation, ultimately leading to dormancy break.
Related News
2025-05-09
2025-05-07
2025-04-29
2025-04-25
2025-04-22
A Comprehensive Overview – Dual-Luciferase Reporter Gene Assay
2025-04-18
2025-04-15
2025-04-10
A Comprehensive Guide to Transcription Factor Research Strategies (Part II)



