A Comprehensive Overview – Dual-Luciferase Reporter Gene Assay
Release time:
2025-04-22
The dual-luciferase reporter gene assay employs Firefly luciferase as the reporter gene and Renilla luciferase as the internal control gene. By simultaneously detecting the activity of both luciferases, this system enables accurate quantitative analysis of target gene expression or molecular interactions.
Principle of Detection
A regulatory element of interest (such as a promoter, miRNA target sequence, etc.) is inserted into an expression vector containing the Firefly luciferase gene, creating a reporter plasmid. This regulatory sequence drives the transcriptional expression of Firefly luciferase. After transfecting the reporter plasmid into cells, the substrate luciferin is added. Firefly luciferase catalyzes the oxidation of luciferin, emitting a luminescent signal (with peak emission around 560 nm). The intensity of this luminescence reflects the activity of the luciferase, allowing for quantitative analysis of gene expression or molecular interactions under different experimental conditions.
Meanwhile, the Renilla luciferase gene is used as an internal control. Both Firefly and Renilla luciferase genes are incorporated into the same plasmid, each driven by a different promoter. This design corrects for variations in
transfection efficiency and transcriptional activity between samples, ensuring the reliability and consistency of the experimental results.

Figure 1: Principle of the Dual-Luciferase Reporter Gene Assay
Main Applications
1.Validation of miRNA–Target Gene Interactions
miRNAs regulate their target genes by inducing mRNA cleavage or translational repression, thereby precisely controlling gene expression levels. By inserting the 3' untranslated region (3'UTR) of the target gene into a luciferase reporter vector, miRNA binding to the inserted sequence leads to the suppression of luciferase translation, resulting in decreased luminescence.
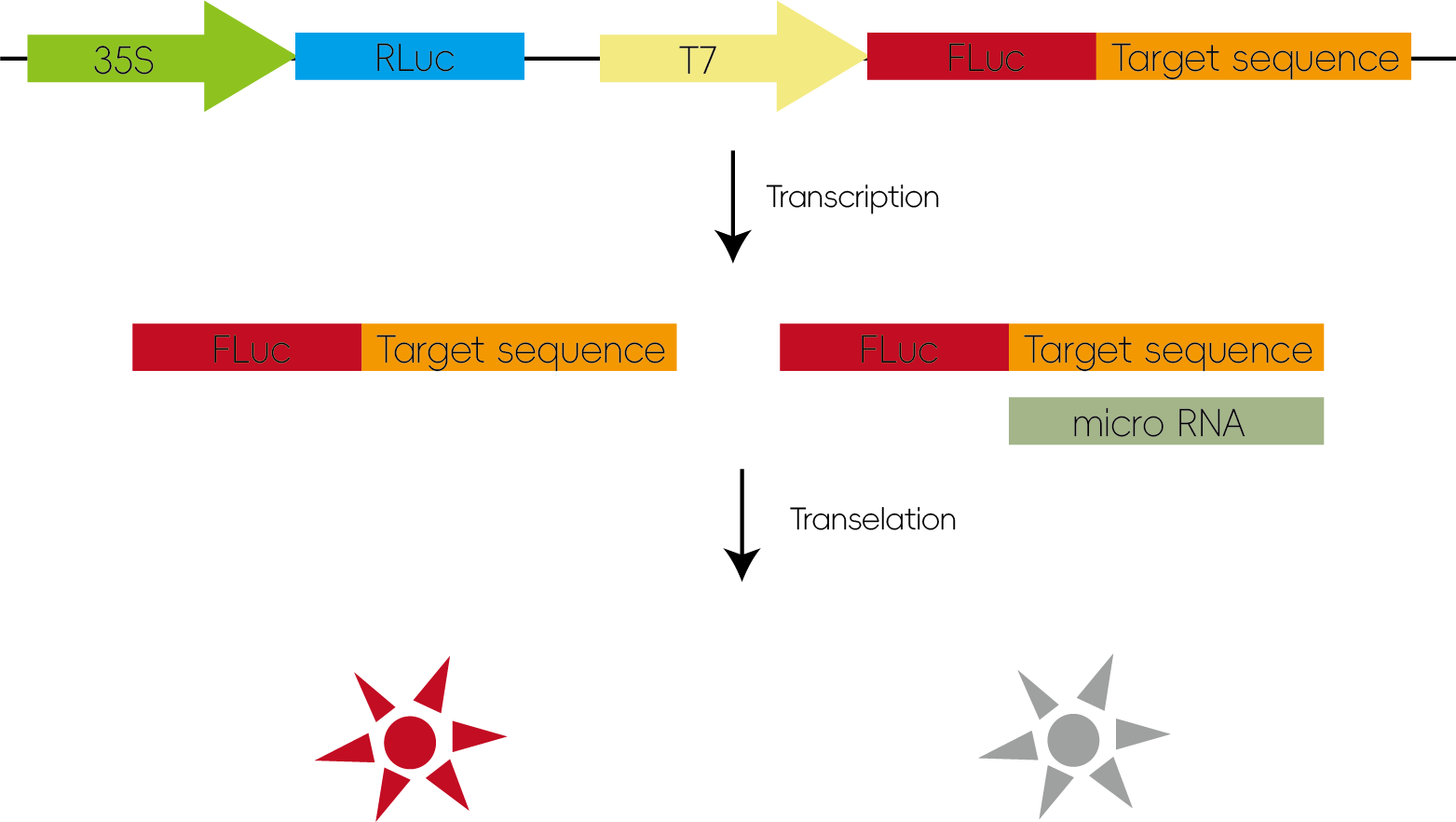
Figure 2: Validation of miRNA–Target Gene Interactions
2.Investigation of miRNA Interactions with lncRNAs or circRNAs
Many lncRNAs have structures similar to mRNAs, and miRNAs can negatively regulate lncRNAs through mechanisms similar to their action on mRNAs. By inserting candidate lncRNA sequences into a luciferase reporter vector, miRNA binding to the inserted sequences can inhibit luciferase translation, thereby reducing luminescence.
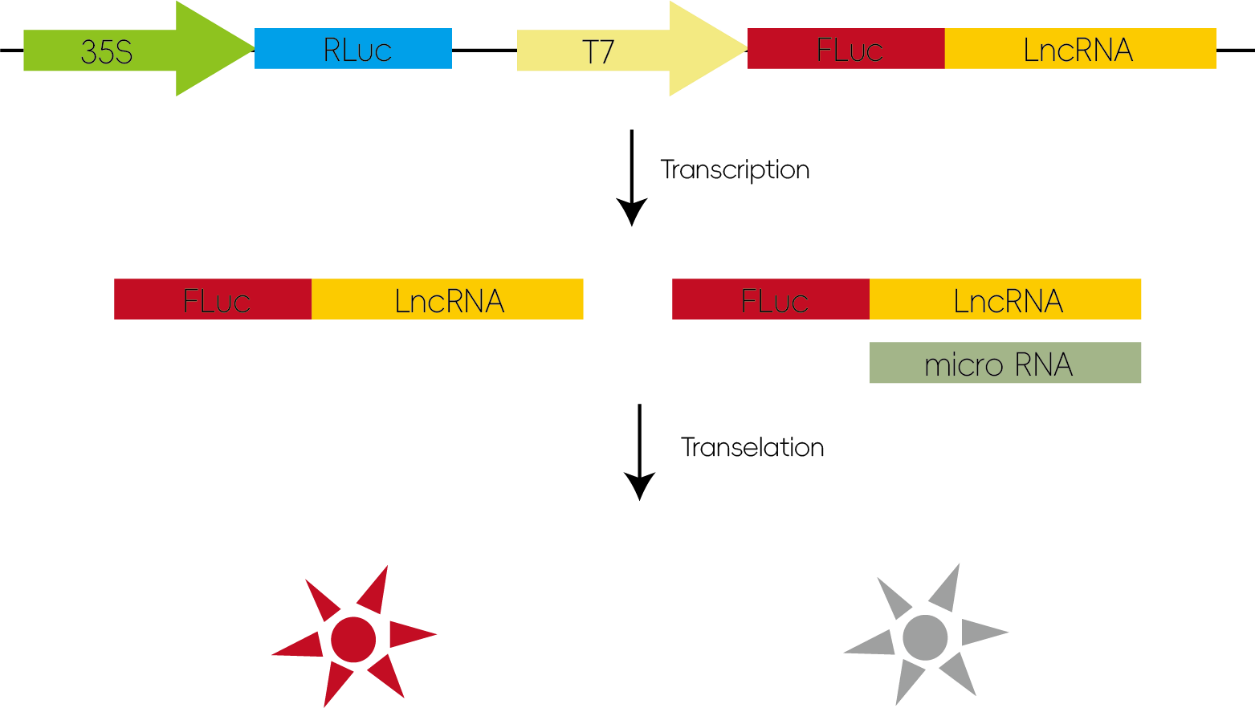
Figure 3: Investigation of miRNA–lncRNA/circRNA Target Interactions
3. Studying the Effects of Transcription Factors on Downstream Genes
Transcription factors regulate gene expression by binding to specific cis-acting elements within the promoter regions of their target genes. When the promoter sequence is inserted upstream of a luciferase gene in a reporter vector and co-transfected with the transcription factor, binding can enhance luciferase expression, leading to increased luminescence.
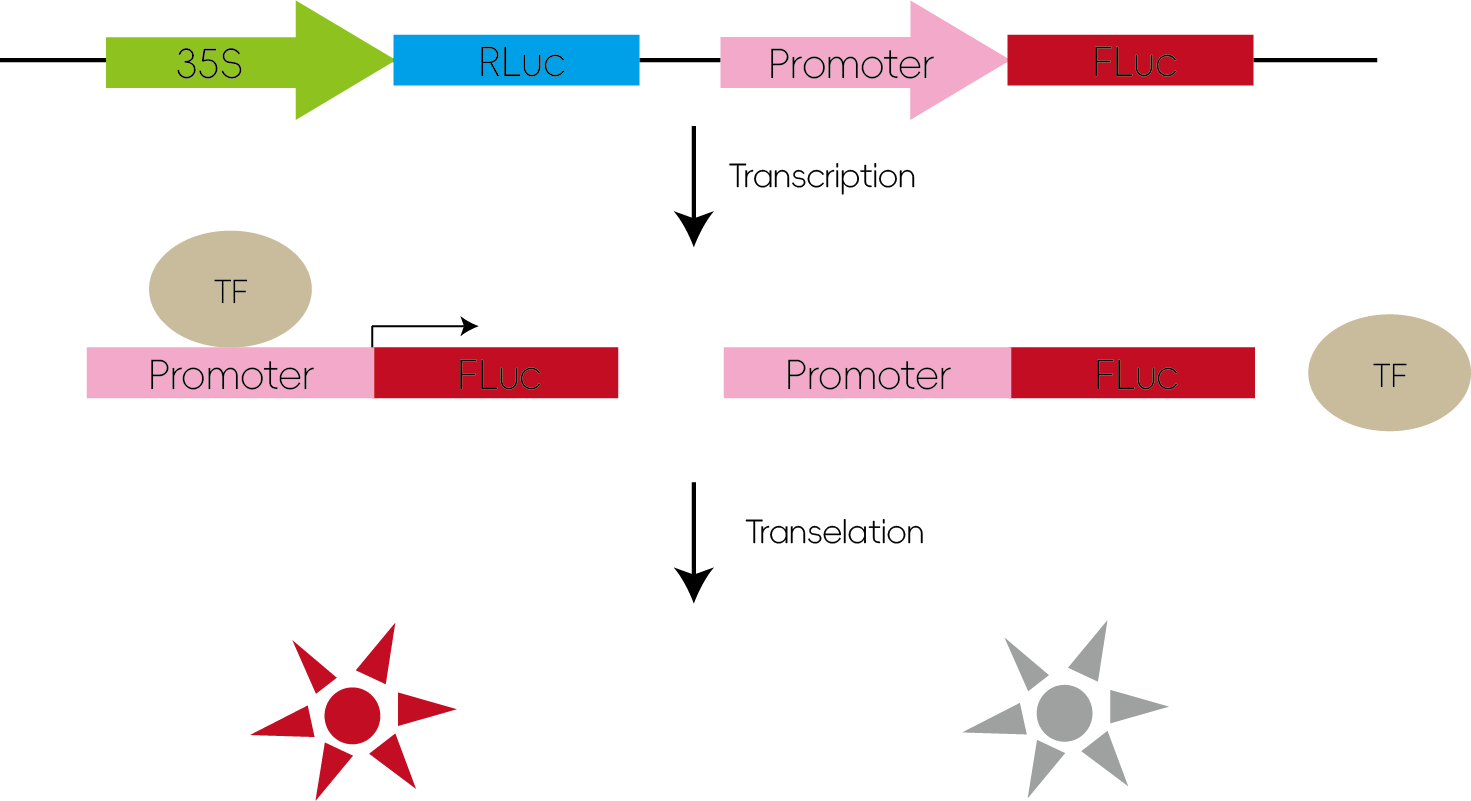
Figure 4: Studying Transcription Factor Regulation of Downstream Genes
4. Promoter Structure Analysis
Promoter regions can be truncated or mutated at specific sites and inserted into luciferase reporter vectors. The resulting constructs are used to assess changes in promoter activity.
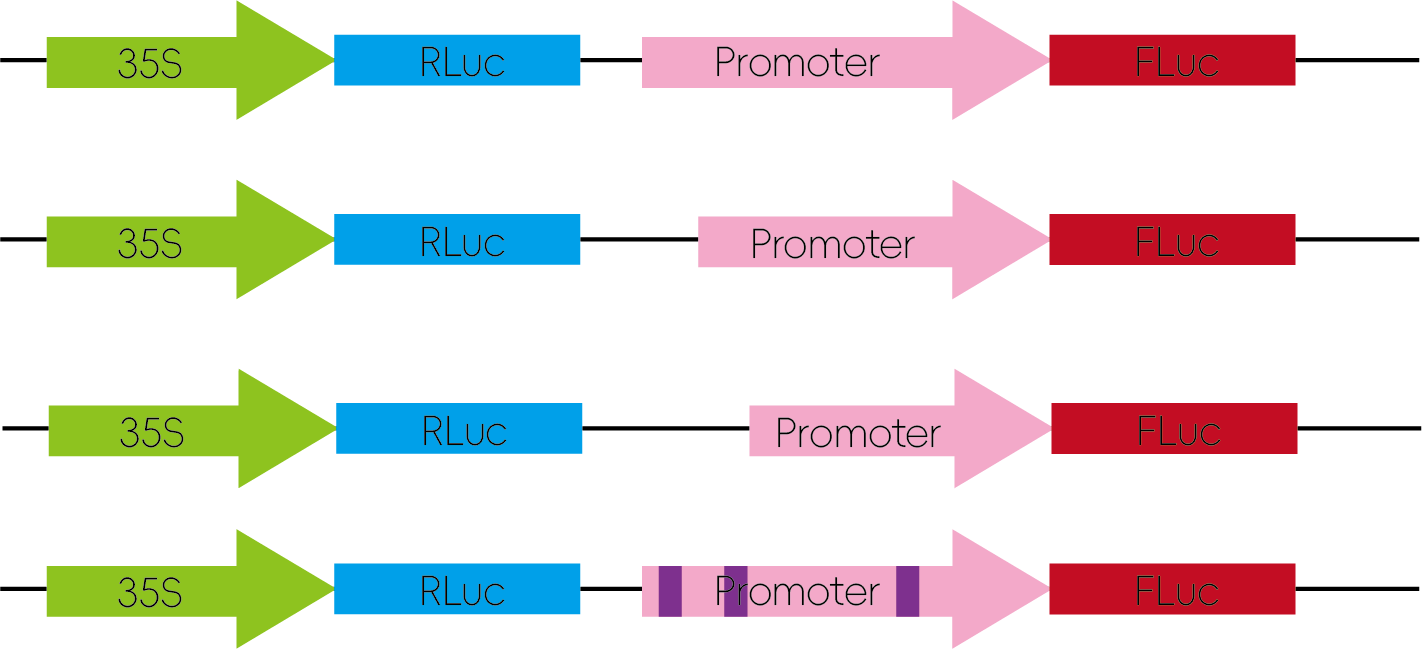
Figure 5: Promoter Structure Analysis
5. Promoter SNP Analysis
Some promoters contain single nucleotide polymorphisms (SNPs). The luciferase reporter system can be used to compare the relative activity of promoter variants.
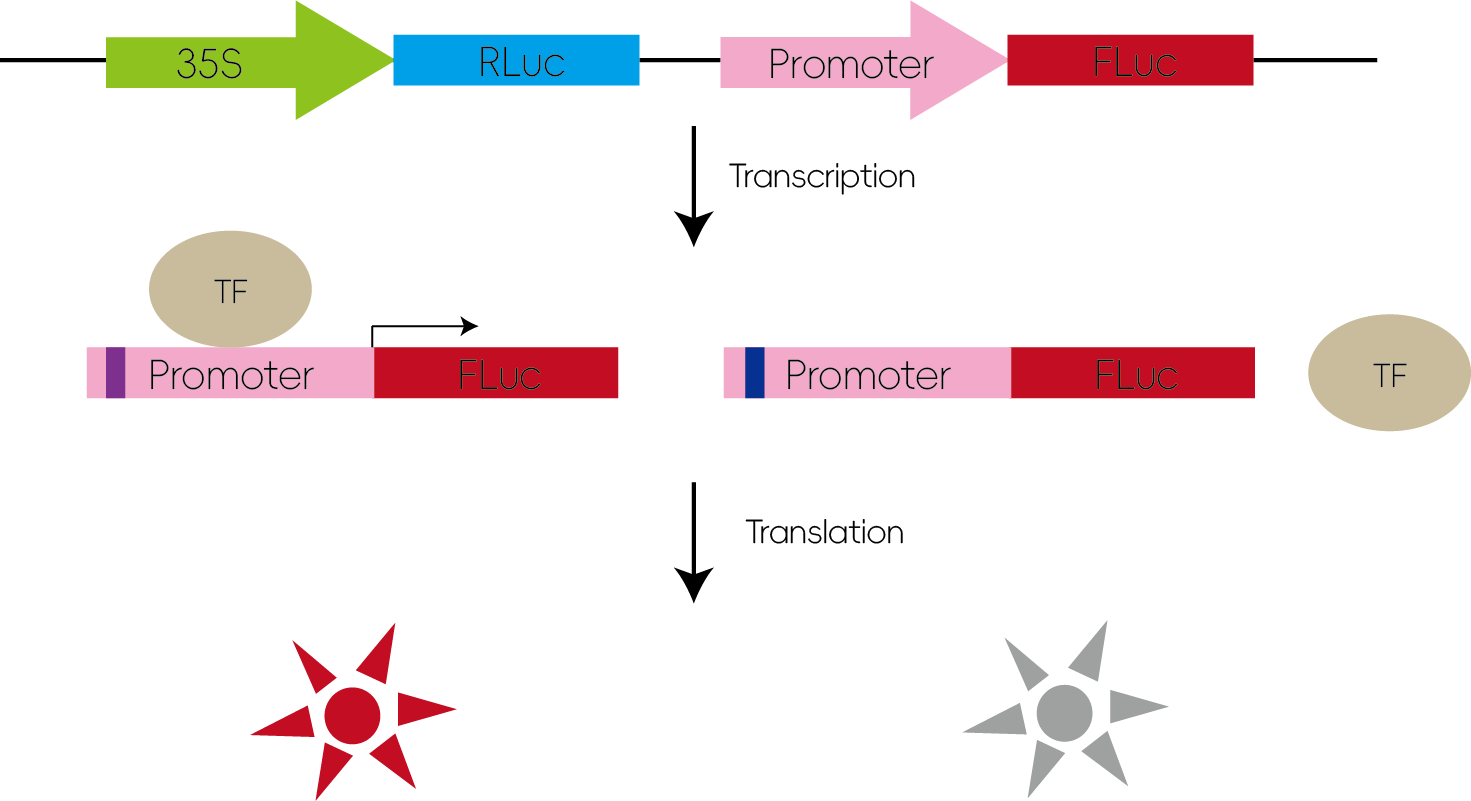
Figure 6: Promoter SNP Analysis
6. Promoter Activity Validation
The promoter of interest is cloned upstream of the luciferase gene in a reporter vector. The resulting luminescence reflects the promoter’s activity.

Figure 7: Promoter Activity Validation
7. Validation of Transcription Factor Activity
The transcription factor of interest is fused with the GAL4 DNA-binding domain and co-transfected with a luciferase reporter construct containing GAL4-responsive elements (GAL-TATA). The luminescence level reflects whether the transcription factor has transactivation or repression activity.
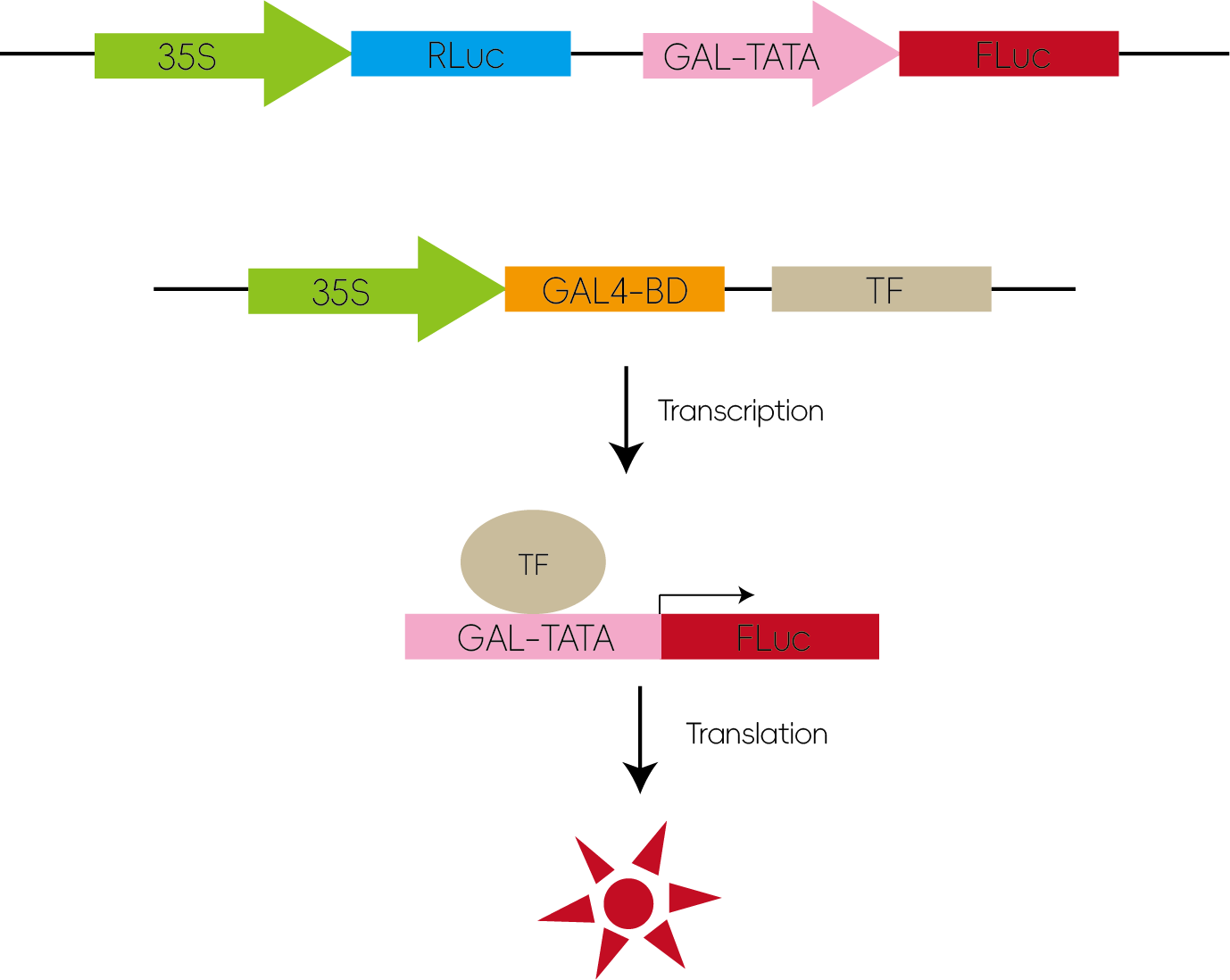
Figure 8: Validation of Transcription Factor Activity
Case Study
On October 7, 2021, a research article titled "ZmMPK5 phosphorylates ZmNAC49 to enhance oxidative stress tolerance in maize" was published in New Phytologist (IF = 8.3). In this study, the authors cloned the promoter sequences of ZmSODs (ZmSOD1, ZmSOD2, ZmSOD3, and ZmSOD4) into the p1381-LUC vector. The effector construct expressed ZmNAC49 under the control of the maize ubiquitin promoter (Ubi:ZmNAC49), with an empty vector used as a control. The results showed that ZmNAC49 activated the LUC expression driven by the ZmSOD3 promoter, indicating that ZmSOD3 is a target gene of ZmNAC49.
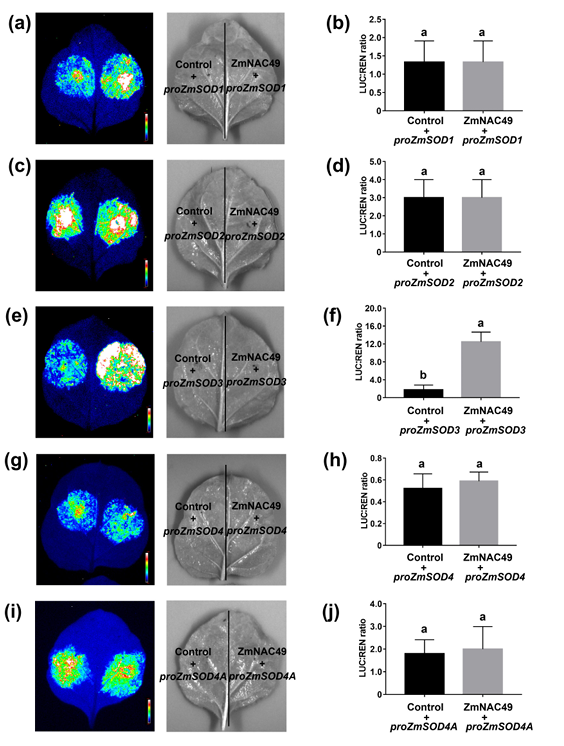
Figure 9: Interaction Study Between ZmSODs and ZmNAC49
In 2020, a research article titled "Novel miR167a-OsARF6-OsAUX3 Module Regulates Grain Length and Weight in Rice" was published in Molecular Plant (IF = 17.1). Through bioinformatics analysis, the authors identified a complementary sequence for miR167a within the coding region of OsARF6. To validate this interaction, they cloned the wild-type 3'UTR of OsARF6 (containing the miR167a binding site) into a luciferase reporter vector, creating pGREEN-OsARF6:LUC.
They also generated a mutant reporter construct, pGREEN-mOsARF6:LUC, by introducing synonymous mutations that disrupted the miR167a binding site. The miR167a precursor was then co-transfected with either the wild-type or mutant reporter into rice protoplasts.
Empty vector ± miR167a precursor (baseline control)
Wild-type OsARF6:LUC ± miR167a precursor
Mutant mOsARF6:LUC ± miR167a precursor
The results showed that the miR167a precursor significantly reduced luciferase activity in the wild-type group, but had no effect in the mutant group, indicating a specific interaction between OsARF6 and miR167a.
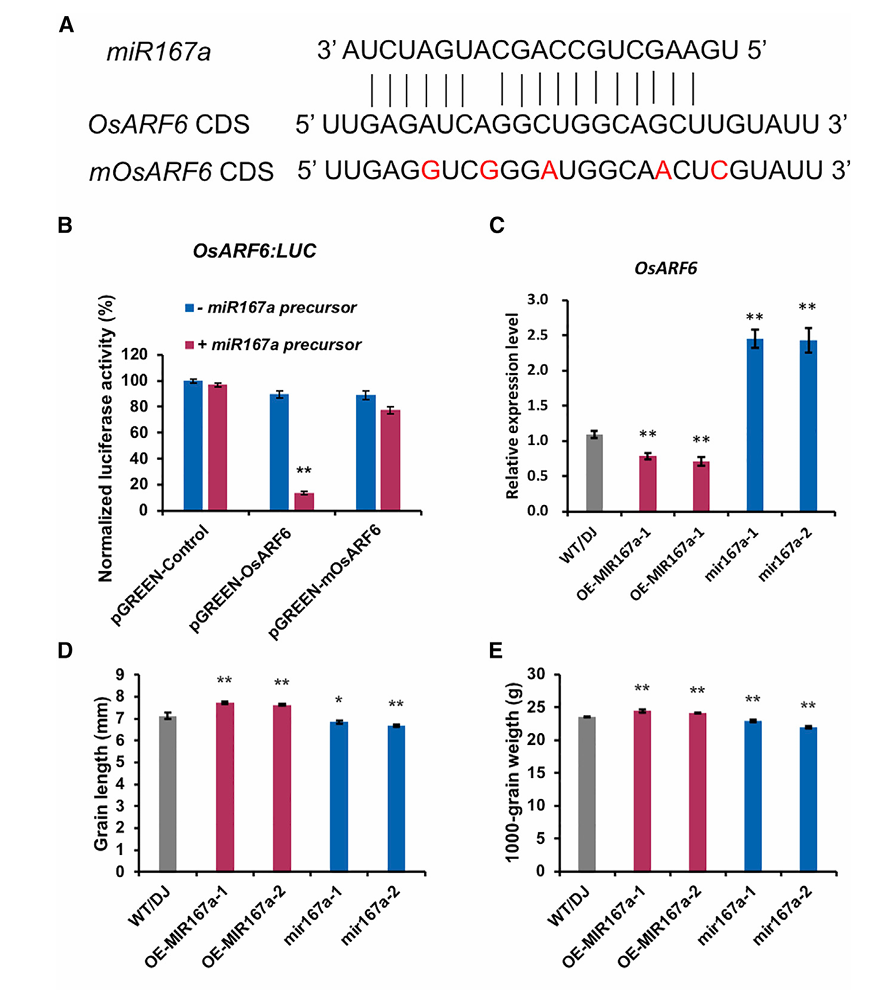
Figure 10: Interaction analysis between OsARF6 and miR167a
Related News
2025-04-25
2025-04-22
A Comprehensive Overview – Dual-Luciferase Reporter Gene Assay
2025-04-18
2025-04-15
2025-04-10
A Comprehensive Guide to Transcription Factor Research Strategies (Part II)
2025-04-08
2025-04-01
A Comprehensive Guide to Transcription Factor Research Strategies (Part 1)
2025-03-27



