Literature Sharing |The LoMYB26/LoJAZ4-LoCOMT module regulates anther dehiscence via Jasmonic acid-mediated endothecium lignification in lily
Release time:
2025-04-15
A working model was proposed in which LoMYB26 interacts with LoJAZ4 to participate in JA-mediated endothecium lignification and anther dehiscence. When bioactive JA (JA-Ile, Me-JA) accumulates, LoJAZ4 is degraded, releasing the transcription factor LoMYB26. LoMYB26 directly binds to the LoCOMT promoter and activates its expression to promote lignin deposition in the anther endothecium. The secondary wall thickening of the anther endothecium results in timely anther dehiscence.

To investigate the variations in lignin content during anther development, lignin accumulation was measured in anthers of S3-S8 stages (Fig. 1A). The results indicated that lignin deposition began at stage S6 and finished at stage S7 (Fig. 1B). The anther endothecium secondary wall thickening occurs from the surrounding cell layers of the anther, generating the required shearing force for anther dehiscence. The histological analysis of normally developed anthers at stages S3-S8 (Fig. 1C) revealed that the endothecium cells formed from stage S3, lignin deposition occurred at stage S6, and the septum degenerated in stages S7-S8. After stages S7-S8, due to septum degradation and the forces generated by lignin deposition, the anther transitioned from four small closed locules to two open locules. However, the pollens did not disperse in these stages. Continuous secondary wall thickening (that fluoresces under UV) of anther endothecium could be observed in stages S3-S8 (Fig. 1D). The anthers at S7 and S8 stages exhibited a higher lignin deposition in the endothecium than the anthers at other stages.
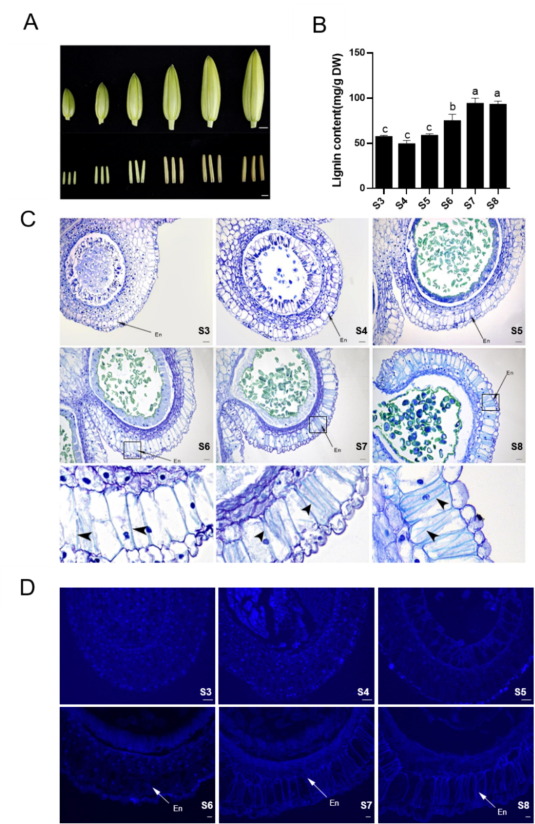
Fig. 1. Lignin accumulation during the anther developmental process.
We used RT-qPCR to detect LoMYB26 expression in the leaf, root, stem, bulbs, petal, anther, stigma, and ovule of lilies (Fig. 2A). LoMYB26 expression was detected in all the organs; however, its accumulation was the highest in the anthers (Fig.2A). Then, we detected the LoMYB26 transcription level in the anthers at S3-S8 stages. LoMYB26 expression was higher in the S6 and S7 stages, which were characterized by lignin deposition and secondary wall thickening (Fig. 2B). Interestingly, LoMYB26 expression was the lowest in the S8 stage, when lignin accumulation in the endothecium was complete. To further determine the effects of LoMYB26 in the anther development, we performed in situ hybridization of LoMYB26 (Fig. 2C). The results indicated that LoMYB26 was expressed in anther endothecial cell layer and middle layer in stages S6 and S7; however, its expression was not detected at stages S5 and S8.
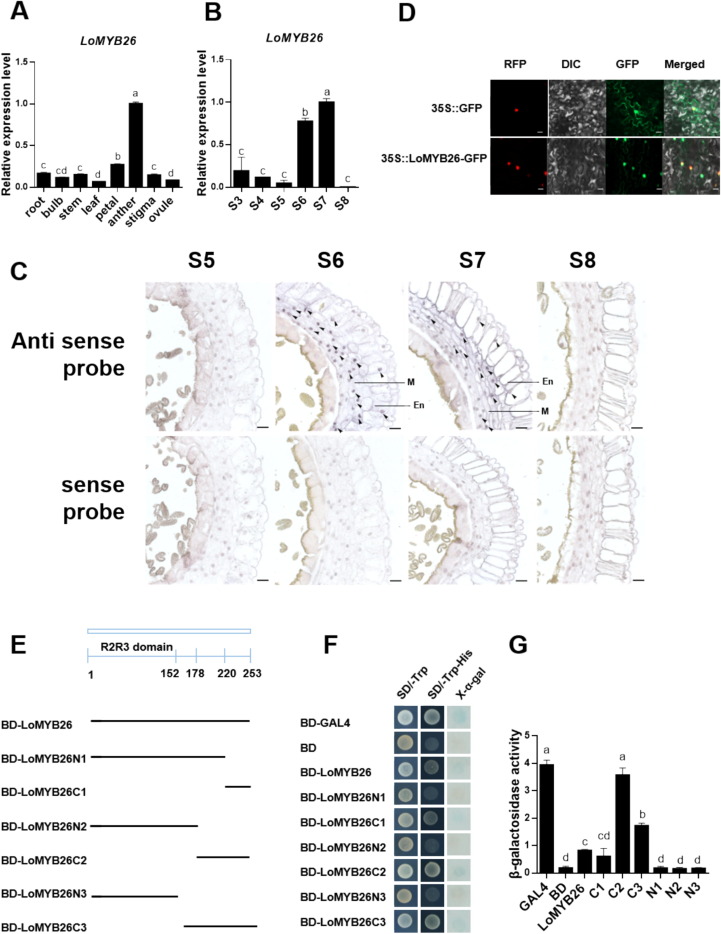
Fig. 2. Analysis of the LoMYB26 expression and transcriptional activation activity.
To further analyze the function of LoMYB26, LoMYB26 was overexpressed in the lily anthers in vitro (Fig. 3A). LoMYB26 expression in the anthers of transiently overexpressing lines was higher than the controls (35S::GFP) (Fig. 3C). The lignin accumulation in 35S::LoMYB26-GFP transiently overexpressing anthers was also higher than that in the 35S::GFP anthers (Fig.3B). The observation of the anther tissue sections revealed that lignin deposition was more pronounced in the endothecium of the 35S::LoMYB26-GFP anthers than the control anthers. The entire endothecium of the overexpressing anthers was filled with lignified fiber bands (Fig. 3D-E); however, the control anthers were covered with lignin only in the surface layer of the cells. These results demonstrated that LoMYB26 overexpression promoted the lignin deposition and the secondary thickening of endothecium. We also observed that the middle layer cells exhibited abnormal swelling and delayed degradation in 35S::LoMYB26-GFP anthers (Fig. 3F). This finding suggested that LoMYB26 might be involved in the development of the middle layer of lily. In addition, the expression of lignin synthase genes LoPAL2, LoCOMT, and Lo4CL2 was higher in 35S::LoMYB26-GFP anthers than the control anthers (Fig. 3C), indicating that LoMYB26 might regulate the expression of lignin synthesis genes to control endothecium lignification.
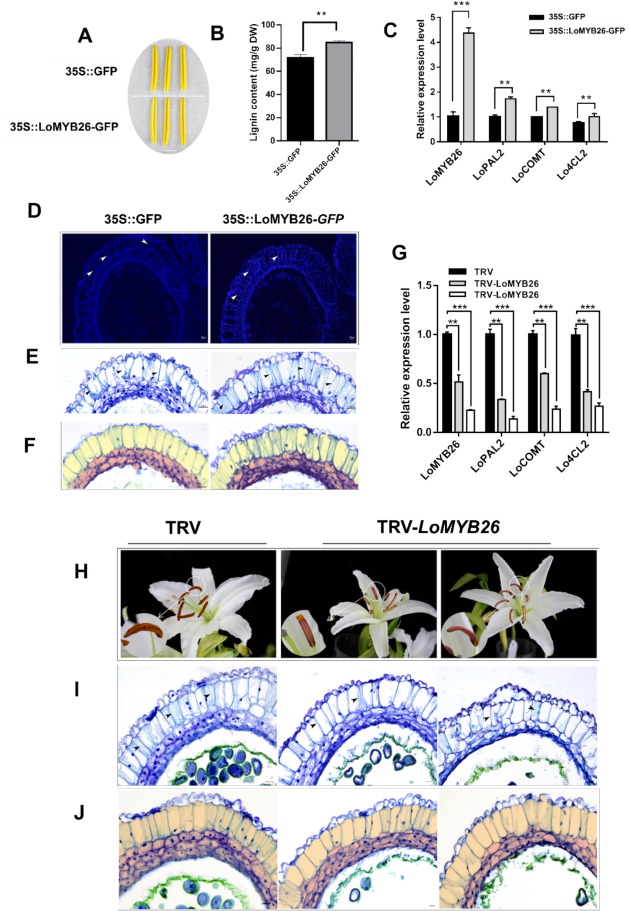
Fig. 3. Lignin formation assay using LoMYB26-overexpressing and LoMYB26-silenced lily anthers.
Since LoMYB26 overexpression activated the expression of LoPAL2, Lo4CL2, and LoCOMT in lily anthers, and LoMYB26 silencing led to opposite effects, we hypothesized that LoMYB26 might directly regulate their expressions. We cloned the promoters of LoPAL2, Lo4CL2, and LoCOMT using the genome of L. davidii var. unicolor as a reference. Y1H assay showed that LoMYB26 bound to the LoCOMT promoter but not to the LoPAL2 and Lo4CL2 promoters (Fig. 4A). EMSA was performed to confirm the interaction of LoCOMT promoter and LoMYB26 in vitro. Our results showed that the His-LoMYB26 protein interacted with the biotin-labeled probes to form shifted signals, and the shifted signals could be diminished by adding the competitive unlabeled probes. However, the control His protein failed to form shifted signals (Fig. 4B). This result indicated that LoMYB26 bound to the cis-element of LoCOMT promoter in vitro. The dual-LUC assay demonstrated that LoMYB26 binds to the LoCOMT promoter and activates it in tobacco leaves (Fig. 4D-E).
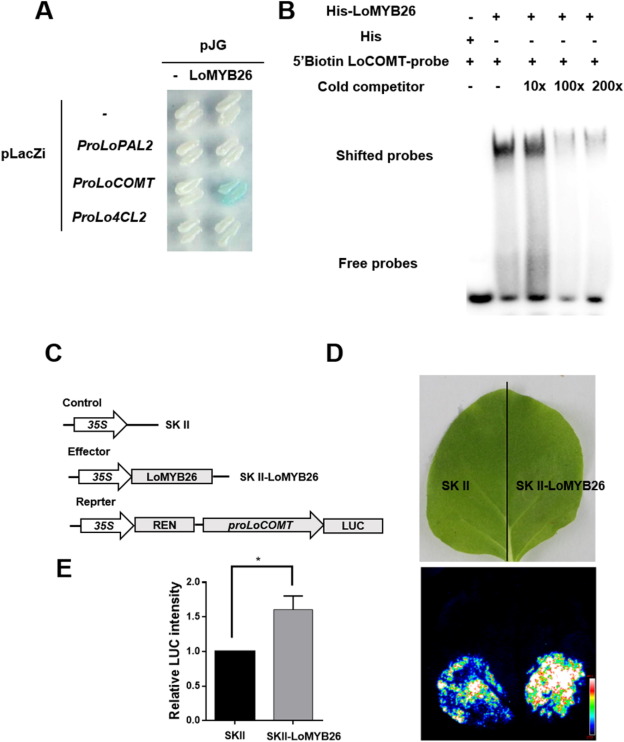
Fig. 4. LoMYB26 activates LoCOMT.
To investigate whether JA affects endothecium lignification, we first conducted JA immunolocalization during the S5-S8 stages of anther development to detect JA distribution in the anther endothecium (Fig. 5A). The fluorescence signals reflecting JA accumulation could be observed in the anther endothecium at S5 and S6 stages, but no signals were detected in the negative control, indicating that JA was accumulated in the endothecium during anther development. These results suggested that JA is involved in the endothecium lignification of lily.
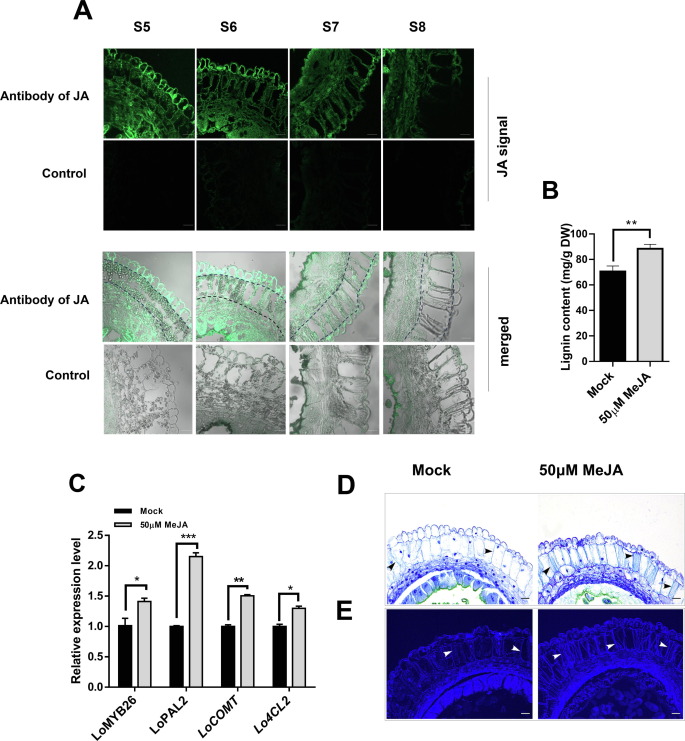
Fig.5. Endothecium lignification after methyl jasmonate (Me-JA) treatment.
To further observe whether Me-JA treatment affects the deposition of lignocellulose in endothecium, we investigated paraffinized sections of anthers at the S7 stage. Toluidine blue staining assay and autofluorescence (under UV light) analysis revealed that lignocellulosic deposits in the Me-JA-treated anther endothecium were more obvious than the Mock (Fig. 5D-E). These results implied that JA-promoted the lignification of the anther endothecium, mediating secondary wall thickening during anther dehiscence by inducing lignin synthase genes.
The gene TRINITY_DN19712_c0_g1_i9_{21} was found to be differentially expressed at the S7 stage of anther development (Fig. S6A). LoMYB26 was differentially expressed in the S5-S7 stages of anther development. These findings implied that TRINITY_DN19712_c0_g1_i9_{21} might interact with LoMYB26. We observed a high LoJAZ4 expression in anthers in the S6 and S7 stages; however, its expression was low in other anther developmental stages (Fig. 6A). In situ hybridization assay was performed to investigate the localization of LoJAZ4 in anthers. Our results showed that LoJAZ4 was expressed in the anther endothecium in the S6 and S7 stages of anther development (Fig. 6B). LoJAZ4 expression was consistent with that of LoMYB26. The subcellular localization assay revealed that LoJAZ4 was located in the nucleus and membrane (Fig. 6C).
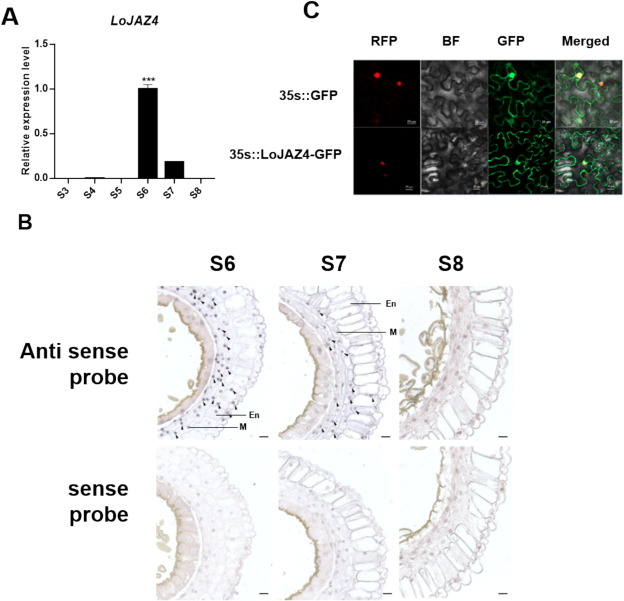
Fig. 6. The expression and subcellular localization of LoJAZ4.
Y2H assays showed that LoMYB26 interacted with LoJAZ4 in yeast (Fig. 7A). LCI assays demonstrated that LoMYB26 interacted with LoJAZ4 in tobacco leaf cells (Fig. 7B). Since JAZ proteins are a class of co-repressors that respond to JA signaling and are typically recruited by the JA receptor coronatine insensitive1 (COI1) during JA signaling, they were subsequently ubiquitinated and degraded to release the repressed transcription factors. We speculated that the interaction between LoMYB26 and LoJAZ4 might be affected by Me-JA treatment, and indeed, this interaction was weakened after treatment with 50 uM Me-JA (Fig. 7C). BiFC assay showed that LoMYB26 interacted with LoJAZ4 in the nucleus (Fig. 7D). Together, these results indicated that LoMYB26 interacted with LoJAZ4 in vitro and in vivo and their interaction was repressed by JA.
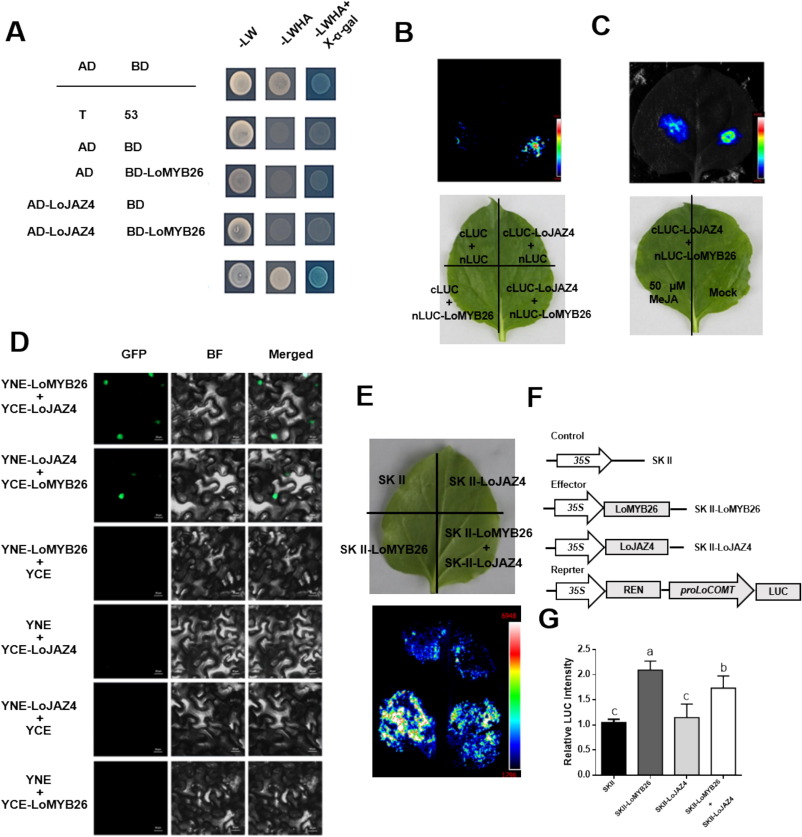
Fig. 7. LoMYB26 interacts with LoJAZ4 in vitro and in vivo.
The dual-LUC assay was performed to determine whether LoJAZ4 contributed to the transactivation ability of LoMYB26 for LoCOMT. In tobacco leaves, it was observed that LUC fluorescence signaling was reduced when LoJAZ4 was co-expressed with LoMYB26, indicating that LoJAZ4 inhibited the transactivation ability of LoMYB26 for LoCOMT (Fig. 7E-G).
Related News
2025-04-25
2025-04-22
A Comprehensive Overview – Dual-Luciferase Reporter Gene Assay
2025-04-18
2025-04-15
2025-04-10
A Comprehensive Guide to Transcription Factor Research Strategies (Part II)
2025-04-08
2025-04-01
A Comprehensive Guide to Transcription Factor Research Strategies (Part 1)
2025-03-27



