Literature Sharing | Aluminum Stress of Oriental Melon (Cucumis melo L.) Is Linked to the Dehydrin CmDHN3
Release time:
2025-05-09
Dehydrins (DHNs), part of the LEA D11 family, are hydrophilic proteins known for their role in plant responses to abiotic stresses, but their functions in cucurbit crops remain largely unexplored. In this study, 34 DHN genes were identified across six cucurbit species, including Cucumis melo. The genes showed high collinearity within subfamilies. Different DHN genes exhibited varying responses to abiotic stresses such as cold, salt, cadmium, and notably aluminum. The acidic SnKn-type gene CmDHN3 in oriental melon (Cucumis melo) was significantly upregulated under aluminum stress. Subcellular localization revealed that CmDHN3 is expressed in the nucleus and cytoplasm. Silencing CmDHN3 led to increased membrane damage and stress indicators (MDA, relative conductivity, and proline), suggesting a protective role under aluminum stress. Transcriptome analysis showed that CmDHN3 downregulation suppressed water channel protein genes. Yeast two-hybrid assays confirmed interactions between CmDHN3 and two aquaporin proteins (CmAQP1 and CmAQP2), suggesting a mechanism of stress response involving water transport. Overall, this study highlights CmDHN3 as a key gene in melon aluminum stress tolerance and provides insight into DHN functions in cucurbits.

Using HMMER-based genome searches with the DHN domain (PF00257), a total of 34 dehydrin (DHN) candidate genes were identified across six cucurbit crops. All identified proteins contained the conserved DHN domain. Specifically, four DHN genes were found in Cucumis sativus, Citrullus lanatus, Cucumis melo, and Cucurbita maxima; five in Lagenaria siceraria; and thirteen in Benincasa hispida.
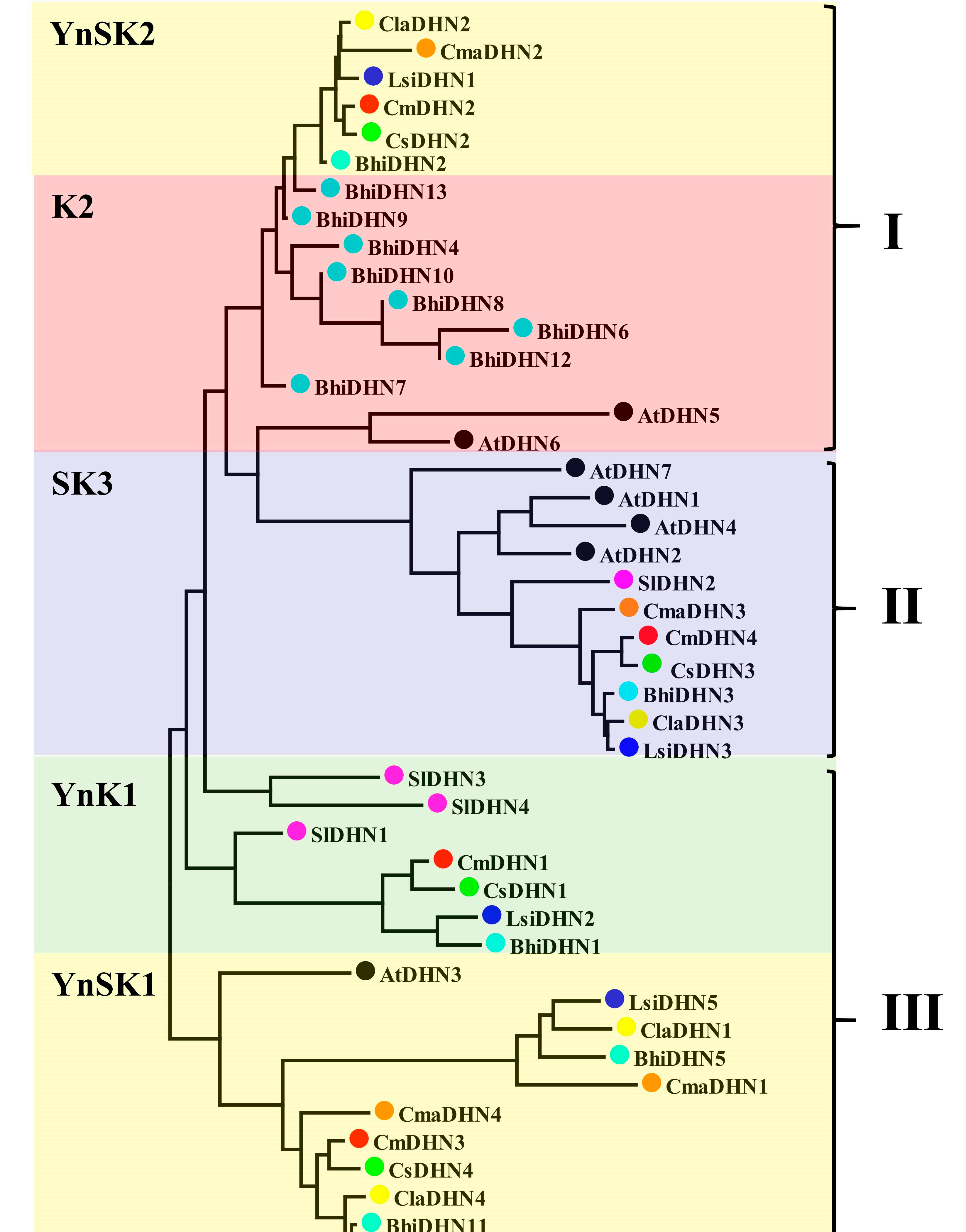
Figure 1. The phylogenetic tree of Cucumis sativus, watermelon, melon, wax gourd, bottle gourd, pumpkin, and Arabidopsis
To explore the evolutionary relationships of DHN genes in cucurbit crops, exon–intron structures of 34 DHN genes were analyzed. The number and arrangement of exons and introns varied notably between subfamilies but were generally conserved within each subfamily.
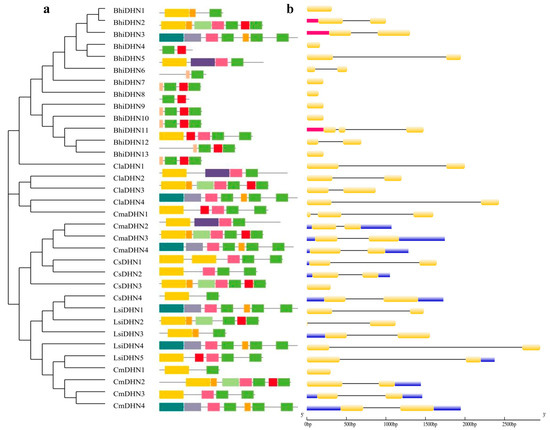
Figure 2 shows (a) conserved motifs of DHN proteins and (b) exon–intron structures in Cucurbitaceae crops. Exons, UTRs, and introns are represented by light blue boxes, yellow boxes, and black lines, respectively
Pairwise collinearity analysis of DHN genes in Cucurbitaceae revealed that closely related species, such as Cucumis sativus and Cucumis melo, had more collinear gene pairs. Collinearity was mainly observed within the same DHN subfamilies, while its absence between subfamilies suggests ancient duplication events gave rise to distinct DHN types.
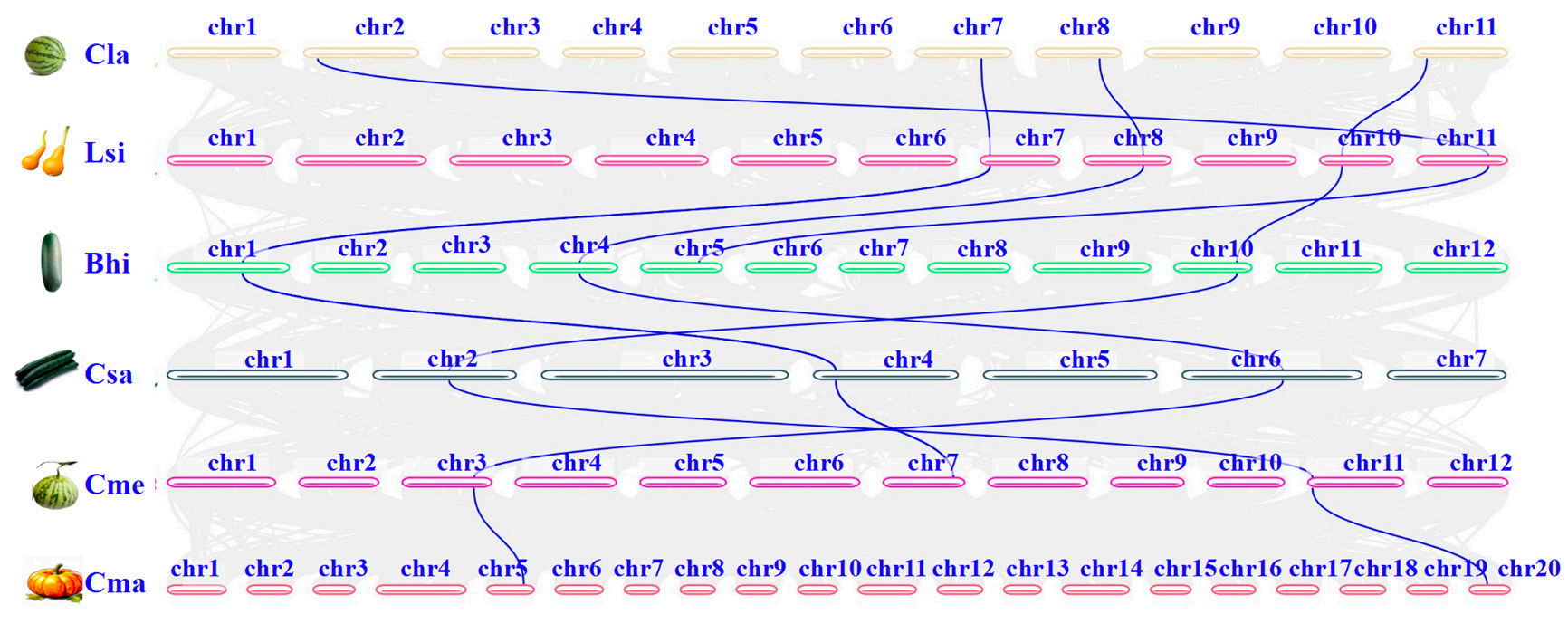
Figure 3. Synteny analysis of DHN genes among the six cucurbit crops.
Expression analysis of DHN genes in six Cucurbitaceae species revealed tissue-specific patterns. While most DHN genes were constitutively expressed and showed high levels in fruits, CmDHN3 and CmDHN4 were predominantly expressed in vegetative tissues. CmDHN2 was highly expressed in all tissues except roots.

Figure 4. The DHN gene expression heatmap of the six cucurbit crops in different tissues.
Expression analysis of Cucumis melo DHN genes under various stresses showed significant variations. Under cadmium (Cd) stress, CmDHN1 was most affected, with a 16-fold increase in roots. Salt stress caused a dramatic increase in CmDHN1 and CmDHN2 expression in both roots (2000-fold and 2100-fold) and leaves (38-fold and 16-fold). Aluminum (Al) stress upregulated CmDHN3 22-fold in roots and 4.8-fold in leaves after 48 hours. Low-temperature stress (4°C) also increased mRNA levels of CmDHN1 and CmDHN2, with a 1300-fold and 2100-fold increase in roots, and a 1800-fold and 2600-fold increase in leaves after 48 hours.
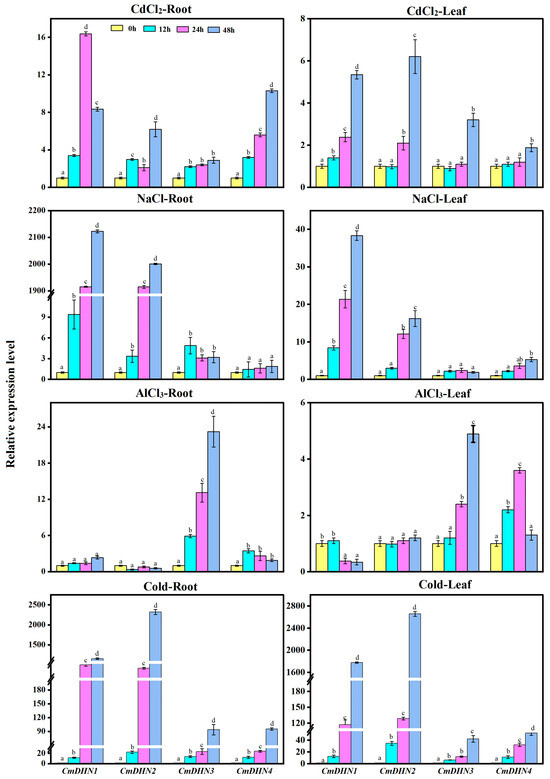
Figure 5. qRT-PCR analysis of CmDHN expression in the roots and leaves of oriental melon plants following abiotic stresses.
To investigate the subcellular localization of CmDHN3 in Cucumis melo, a GFP-tagged vector (pBI221-CmDHN3-GFP) was constructed and transformed into Arabidopsis protoplasts. The expression site of CmDHN3 was identified using green fluorescent protein (GFP) signals, revealing that CmDHN3 is expressed in both the nucleus and cytoplasm.
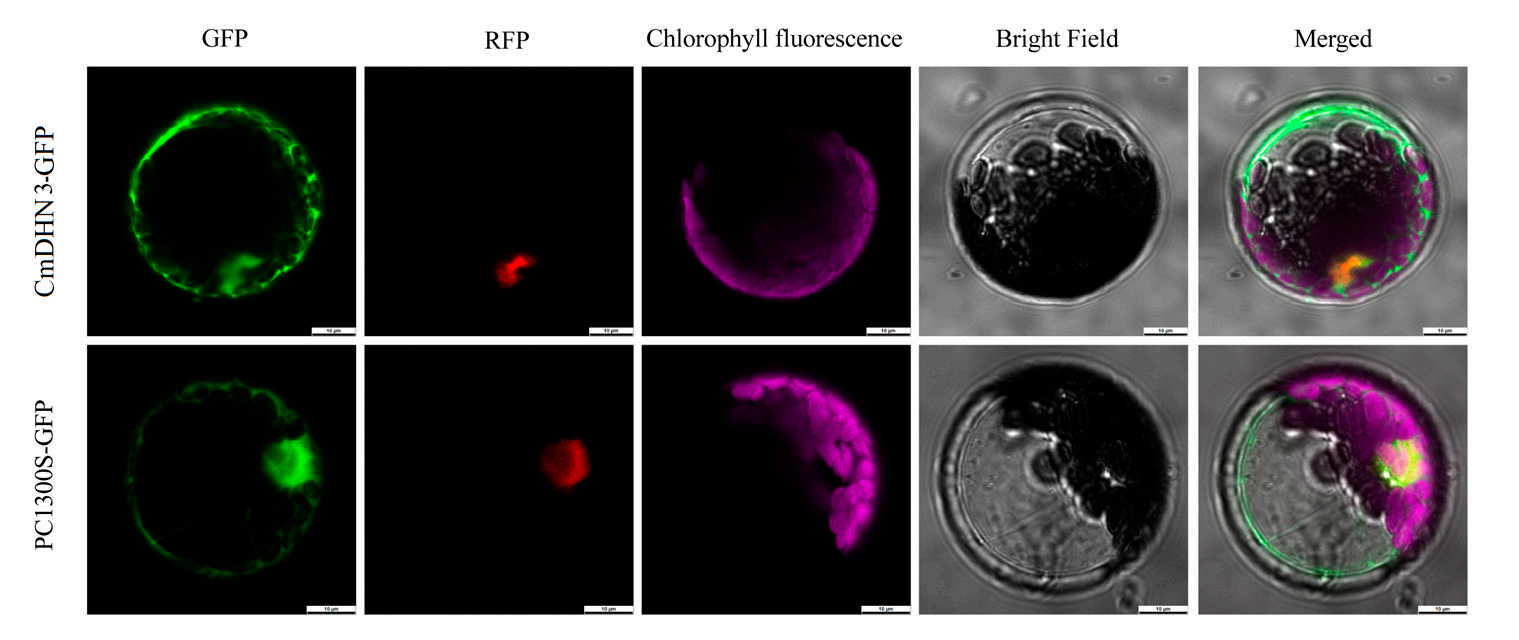
Figure 6. The subcellular localization of Cucumis melo CmDHN3 in Arabidopsis leaf protoplasts.
To explore the role of CmDHN3 in aluminum (Al) stress response, a VIGS (virus-induced gene silencing) approach was used in Cucumis melo. The CmDHN3 gene was silenced using the pTRV2-CmDHN3 vector. No visible phenotypic differences were observed in the silenced plants under normal conditions. However, under Al stress, CmDHN3-silenced plants showed significant growth inhibition and root system suppression compared to the control. Additionally, CmDHN3 expression in the roots and leaves of silenced plants was significantly lower than in controls, confirming the silencing efficiency.
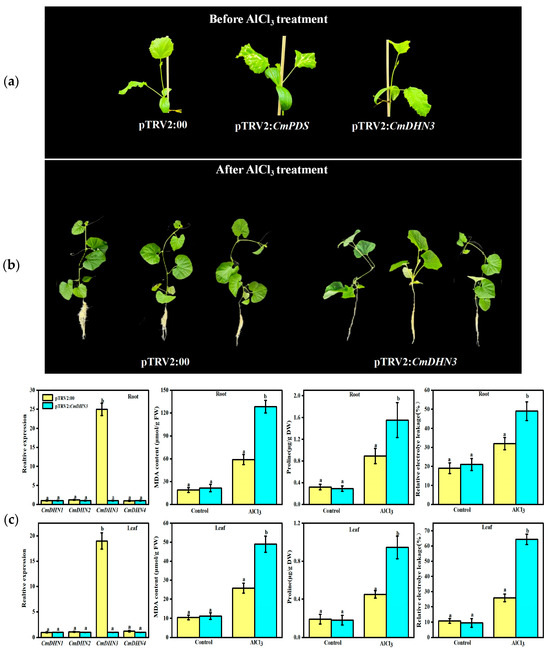
Figure 7. Effects of Al stress on plant phenotypes
Under aluminum (Al) stress, CmDHN3-silenced plants showed higher malondialdehyde (MDA) and proline content, and greater relative conductivity, compared to control plants. Specifically, MDA levels were 2.3-fold and 1.9-fold higher in the roots and leaves of silenced plants, respectively, while proline content increased by 1.4-fold and 2.1-fold. After two weeks of Al stress, transcriptomic analysis revealed 1284 differentially expressed genes (DEGs), with 575 downregulated and 708 upregulated. KEGG analysis showed these DEGs were enriched in pathways like MAPK signaling, hormone signaling, and glutathione metabolism. Key genes such as CmAQP1, CmAQP2, and CmZIP were significantly downregulated in CmDHN3-silenced plants.
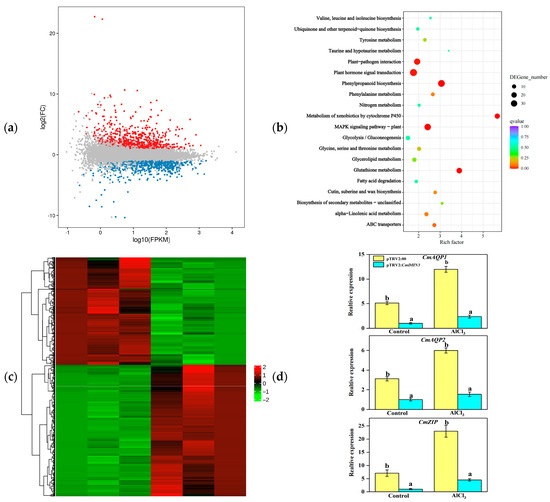
Figure 8. Overview of CmDHN3-silenced Cucumis melo plants transcriptomes of roots under Al stress on day 14

Figure 9. Validation of the protein that interacts with CmDHN3.
This study identified 34 DHN genes in the Cucurbitaceae genome, with varying expression patterns in different tissues and under stress conditions. The acidic SnKn-type DHN gene CmDHN3 was upregulated under Al stress, and its silencing decreased Al stress tolerance in melon. Additionally, silencing CmDHN3 reduced the expression of water channel proteins CmAQP1 and CmAQP2, as well as the ABA signaling factor CmZIP. Y2H assays confirmed interactions between these proteins. The findings provide insights into the role of CmDHN3 in Al stress response in melons.
Related News
2025-05-13
2025-05-09
2025-05-07
2025-04-29
2025-04-25
2025-04-22
A Comprehensive Overview – Dual-Luciferase Reporter Gene Assay
2025-04-18
2025-04-15



