Literature Sharing | SoNAC72-SoMYB44/SobHLH130 module contributes to flower color fading via regulating anthocyanin biosynthesis by directly binding to the SoUFGT1 promoter in lilac (Syringa oblata)
Release time:
2025-05-22
This study reveals that flower color fading in lilac (Syringa oblata) is caused by decreased anthocyanin accumulation during flower development. The enzyme SoUFGT1, essential for stabilizing anthocyanins, is regulated by three transcription factors: SoMYB44, SobHLH130, and SoNAC72. SoMYB44 activates SoUFGT1 expression, while SoNAC72 promotes SoMYB44 by binding to its promoter. SobHLH130 interacts with SoMYB44 to enhance its regulatory effect. As flowers age, reduced expression of SoNAC72 and SobHLH130 lowers SoMYB44 levels, suppressing SoUFGT1 expression and anthocyanin biosynthesis, leading to flower color fading.

To investigate the cause of petal color fading in lilac, anthocyanin levels were measured across flower development stages. The results showed a 60% decrease in anthocyanin content from the bud stage (S1) to full bloom (S2), correlating with visible petal fading. Transcriptome analysis between S1 and S2 stages identified differentially expressed genes, among which SoUFGT1 showed an expression pattern consistent with anthocyanin changes, suggesting its key role in anthocyanin biosynthesis. The 1146 bp coding sequence of SoUFGT1 was cloned, and bioinformatics analysis revealed it encodes a stable, hydrophilic protein highly conserved among plant species. qRT-PCR confirmed its expression pattern, with highest levels in roots and flowers at S2. Subcellular localization showed that SoUFGT1 is located in the cytoplasm. These findings support SoUFGT1 as a pivotal gene in regulating anthocyanin accumulation in lilac petals.
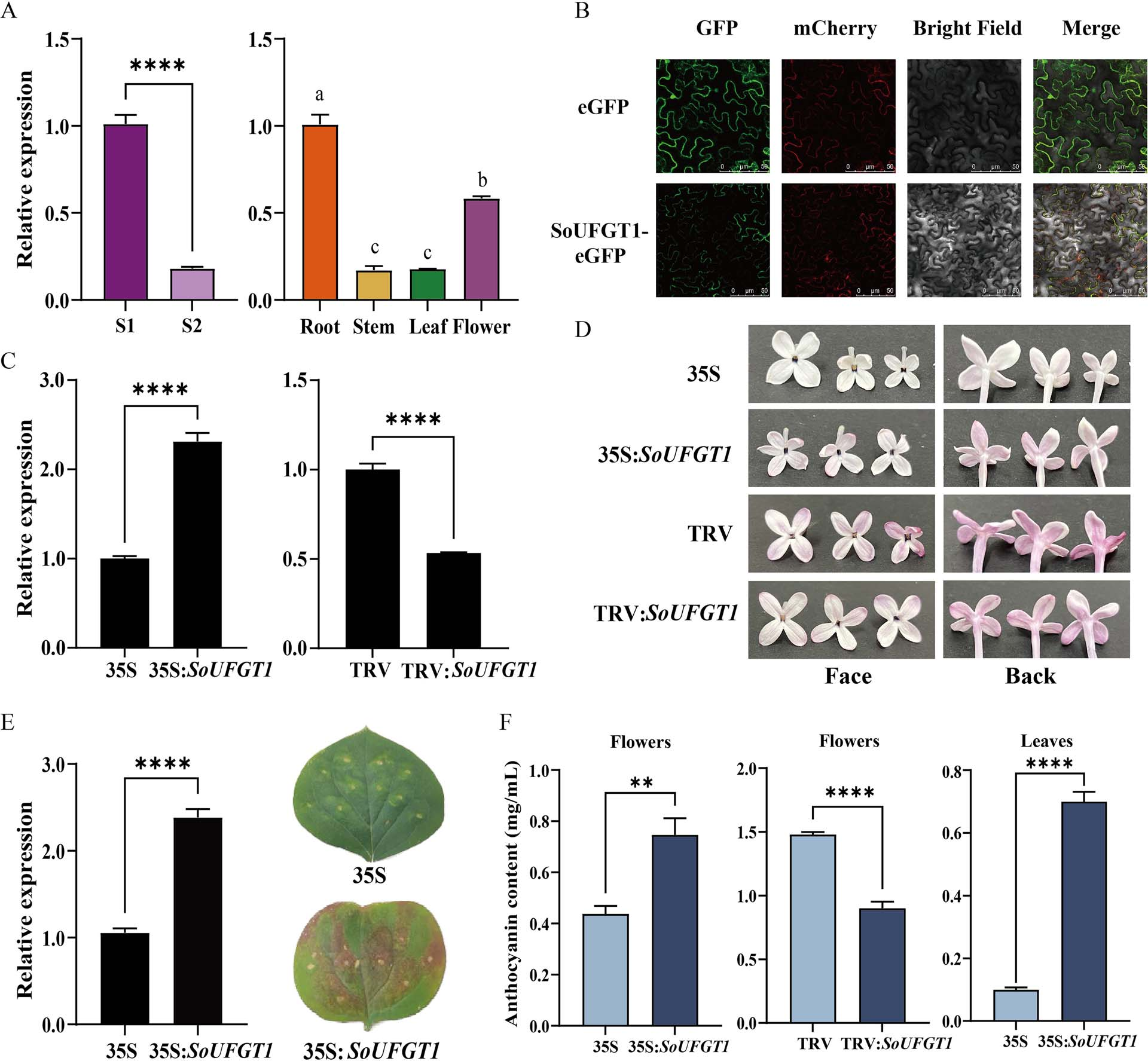
Figure 2. SoUFGT1 expression pattern, subcellular localization, and functional analysis
Using time-ordered gene co-expression networks and transcriptome data from S1 and S2 stages, a MYB transcription factor associated with SoUFGT1 expression was identified and named SoMYB44. Its expression was higher at S1 than S2, mirroring the patterns of SoUFGT1 and anthocyanin content. SoMYB44 also showed high expression in roots and flowers at S2. The 1023 bp full-length coding sequence was cloned, and phylogenetic analysis revealed close similarity to a homolog in Olea europaea. Subcellular localization indicated that SoMYB44 functions as a nuclear transcription factor.
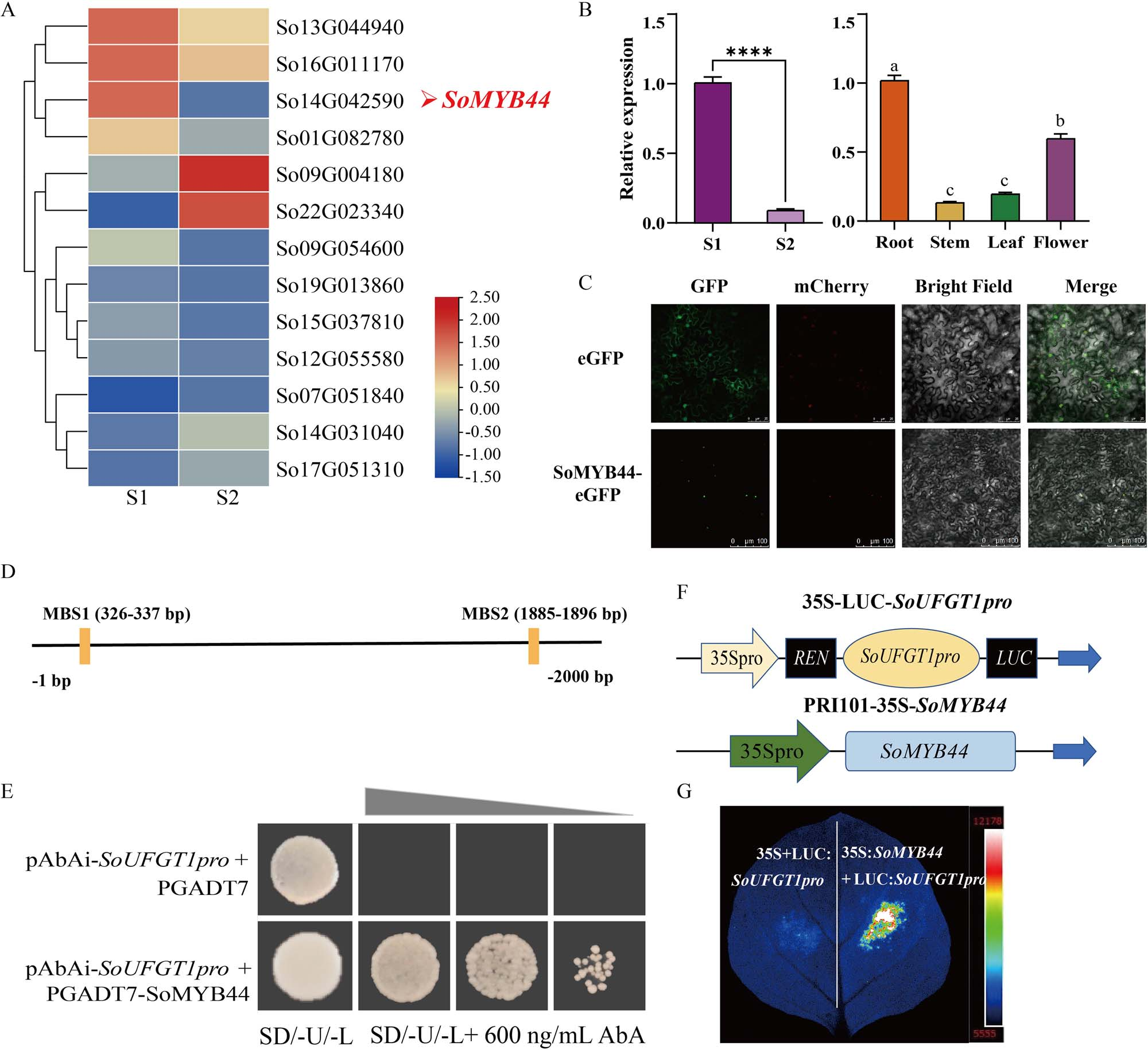
Figure 3. Relationship between SoMYB44 and SoUFGT1
Given its expression pattern, SoMYB44 was hypothesized to positively regulate anthocyanin biosynthesis. To test this, SoMYB44 was transiently overexpressed and silenced in lilac petals. qRT-PCR confirmed successful manipulation, with overexpression increasing SoMYB44 levels by 2.8-fold and silencing reducing it by ~49%. Phenotypically, overexpression darkened petal color and increased anthocyanin content to nearly double that of the control, while silencing led to lighter petals and reduced anthocyanin levels. In leaves, overexpression also enhanced pigmentation, resulting in a 5-fold increase in anthocyanin content. These results support SoMYB44 as a positive regulator of anthocyanin biosynthesis.
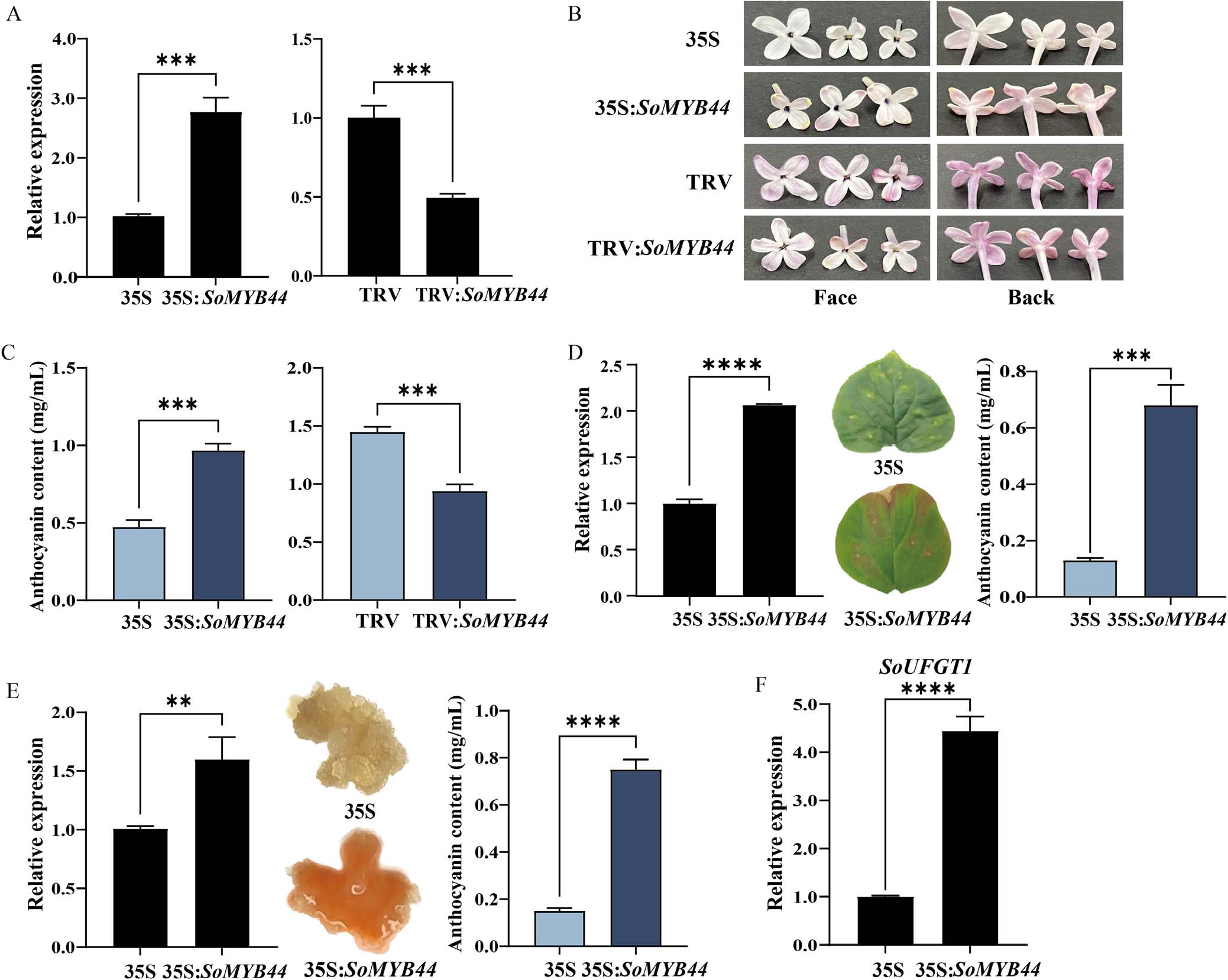
Figure 4. The function analysis of SoMYB44
To clarify how SoMYB44 regulates anthocyanin accumulation, its potential interacting partners SobHLH130 and SoNAC72 were identified from transcriptome data and validated by qRT-PCR. Both genes showed higher expression at the S1 stage. Functional analysis revealed that overexpression of either gene darkened petal color and doubled anthocyanin content, while silencing led to lighter petals and a ~33% reduction in anthocyanins. Similar effects were observed in leaves and calli, confirming that SobHLH130 and SoNAC72 are positive regulators of anthocyanin biosynthesis in lilac.
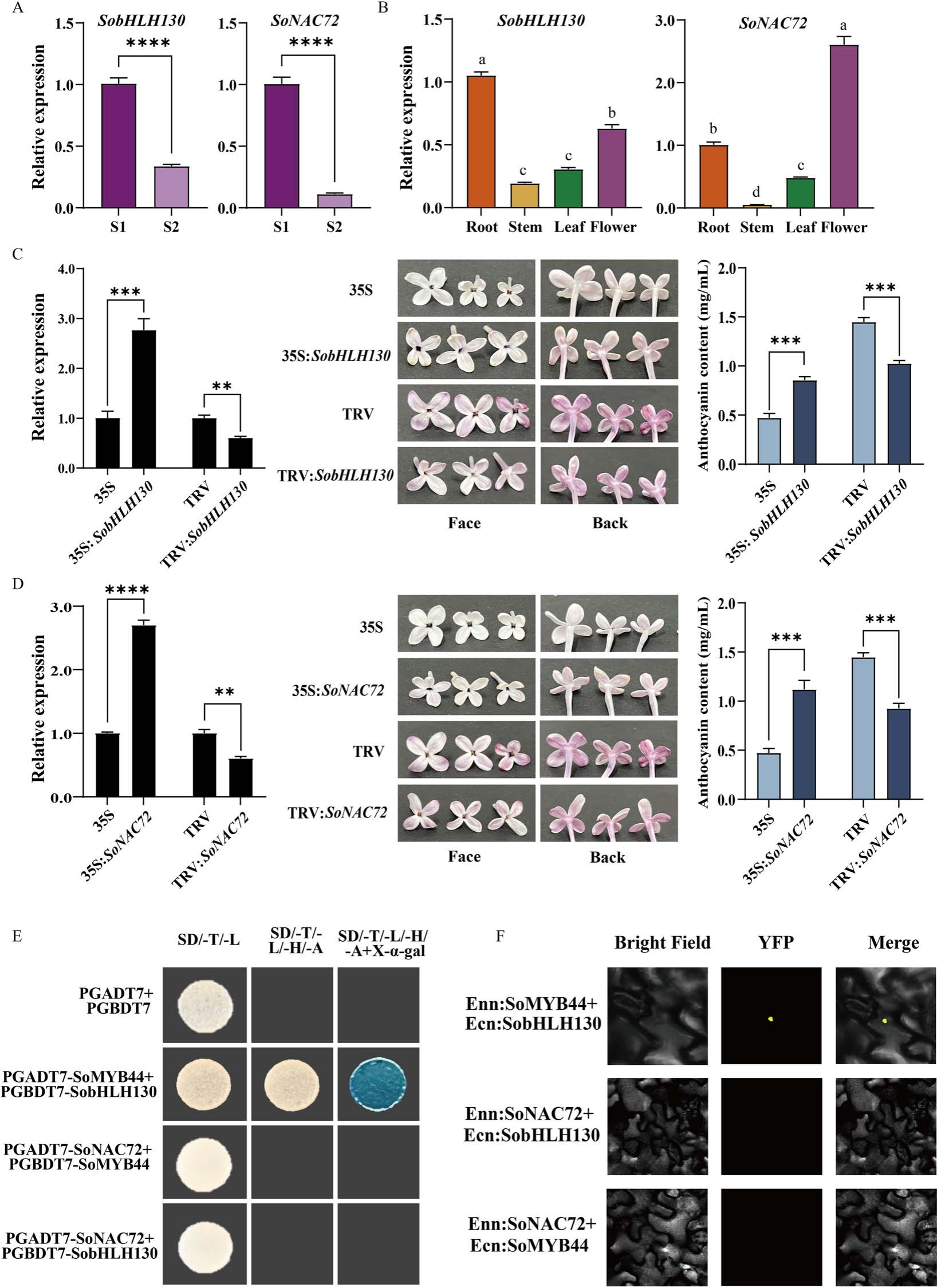
Figure 5. Function of SobHL H130 and SoNAC72, and relationship with SoMYB44
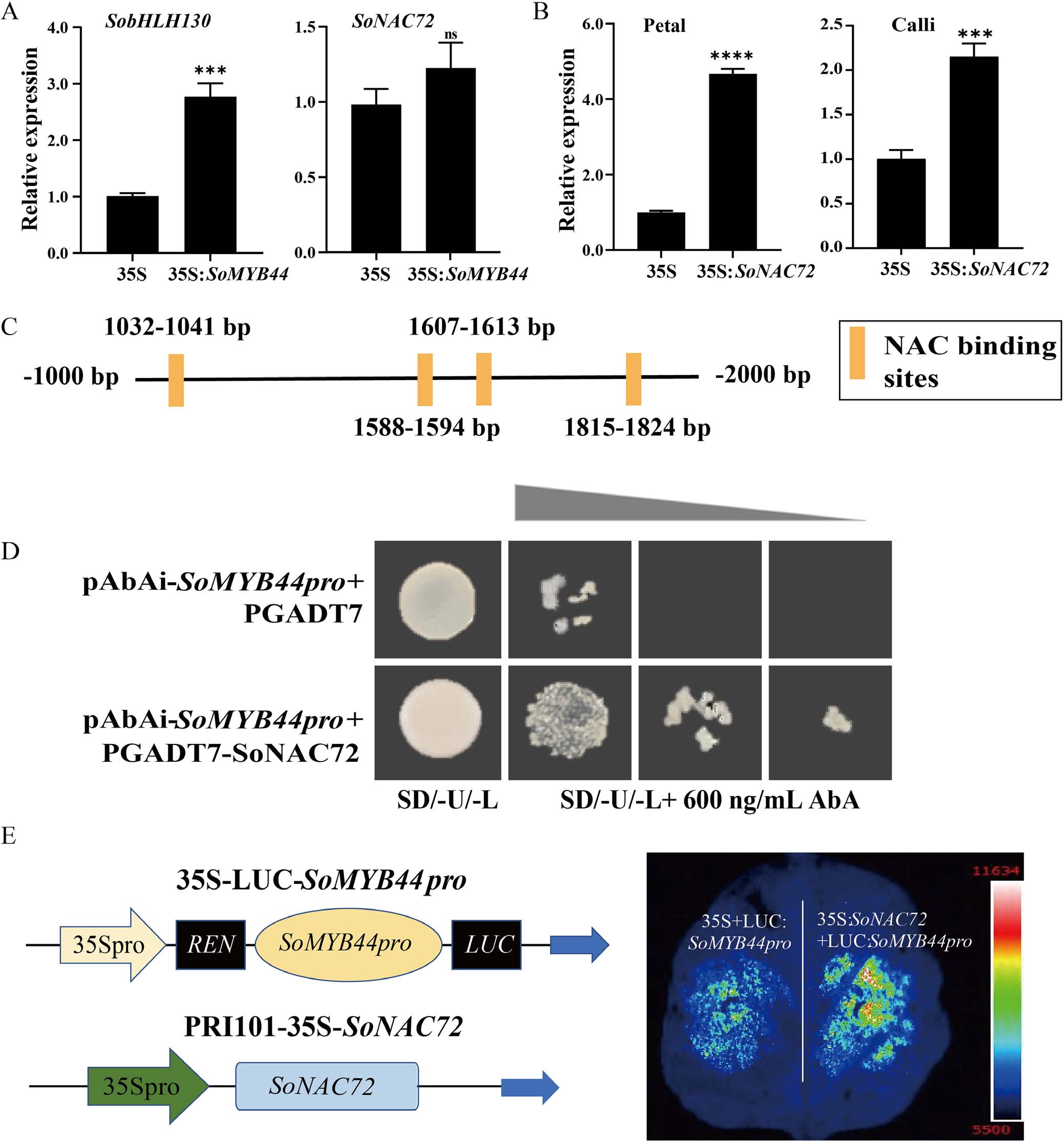
Figure 6. Relationship among SoMYB44 and SoNAC72
In lilac petals overexpressing SoMYB44, SobHLH130 expression increased significantly, while SoNAC72 did not. However, overexpression of SoNAC72 elevated SoMYB44 expression in both petals and calli, suggesting that SoNAC72 may regulate SoMYB44. Promoter analysis revealed NAC binding sites in the SoMYB44 promoter region. Yeast one-hybrid and dual-luciferase assays confirmed that SoNAC72 directly binds to and activates the SoMYB44 promoter, indicating that SoNAC72 functions upstream of SoMYB44 in regulating anthocyanin biosynthesis.
In summary, the SoNAC72–SoMYB44/SobHLH130 module regulates anthocyanin biosynthesis in lilac petals by activating SoUFGT1 expression, with high transcription factor activity in early flower stages leading to darker color. As flowers mature, decreased expression of these TFs weakens SoUFGT1 activation, reducing anthocyanin accumulation and causing color fading. These findings help explain lilac petal color fading and provide candidate genes for floral color improvement.
Related News
2025-06-04
Functional study of plant hormone transporters using a heterologous system
2025-05-29
2025-05-27
2025-05-22
2025-05-20
2025-05-16
2025-05-13
2025-05-09



