Literature Sharing | Analysis of the Transcriptome Provides Insights into the Photosynthate of Maize Response to Salt Stress by 5-Aminolevulinic Acid
Release time:
2025-05-29
Salt stress significantly hinders maize growth and yield. This study explored the role of exogenous 5-aminolevulinic acid (ALA) in regulating photosynthesis in maize seedlings under salt stress. Transcriptome and physiological analyses of the “Zhengdan 958” cultivar revealed 4634 differentially expressed genes, including key transcription factors (NAC, MYB, WRKY). These genes were enriched in pathways related to porphyrin metabolism, photosynthesis, and carbon fixation. ALA treatment enhanced photosynthetic gene expression, pigment levels, antioxidant enzyme activities (SOD and CAT), reduced ROS accumulation, and increased starch content. Overall, ALA improves maize salt tolerance by modulating photosynthesis-related pathways.

Salt stress significantly affected the phenotypic and anatomical characteristics of maize leaves. After 72 hours of 300 mmol/L NaCl treatment, seedlings showed stunted growth, smaller leaves, and severe wilting. Exogenous ALA application alleviated these effects and promoted better growth. Microscopic analysis revealed that control plants had well-organized leaf structures, including clearly defined bundle sheath cells (BSCs) and numerous chloroplasts. Salt stress disrupted this structure, resulting in disorganized BSCs and fewer, irregular chloroplasts. However, ALA treatment under salt stress preserved leaf structure, improved BSC definition, and increased the number and organization of chloroplasts.
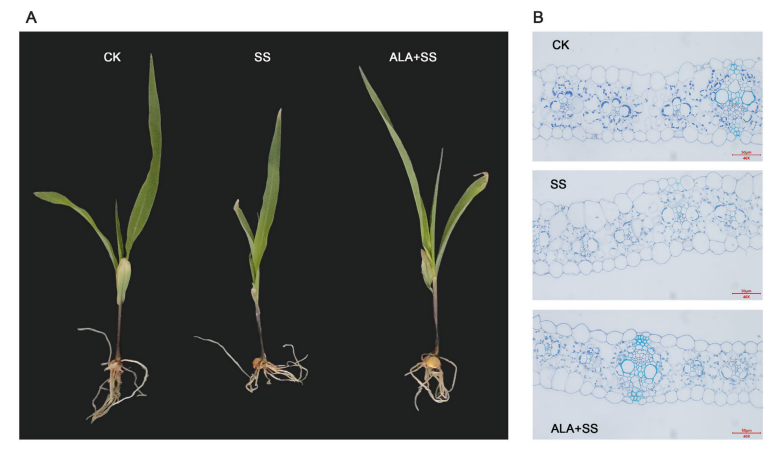
Figure 1. Effect of ALA on (A) phenotype and (B) microstructure of maize leaves under salt stress
To examine ALA-regulated gene expression under salt stress, transcriptome sequencing was performed on maize leaf samples from three treatment groups (CK, SS, and ALA_SS), each with three biological replicates. The sequencing generated over 413 million high-quality reads, with an average mapping rate of 88.49% to the maize reference genome. Quality metrics (Q20 > 97.26%, Q30 > 94.98%) confirmed the data were suitable for downstream analysis. A total of 21,011, 19,628, and 20,724 genes were identified in CK, SS, and ALA_SS groups, respectively, with 17,525 genes shared among all groups. Principal component analysis (PCA) and correlation analysis showed clear separation between treatment groups and high consistency among replicates, supporting the reliability of the experimental design.
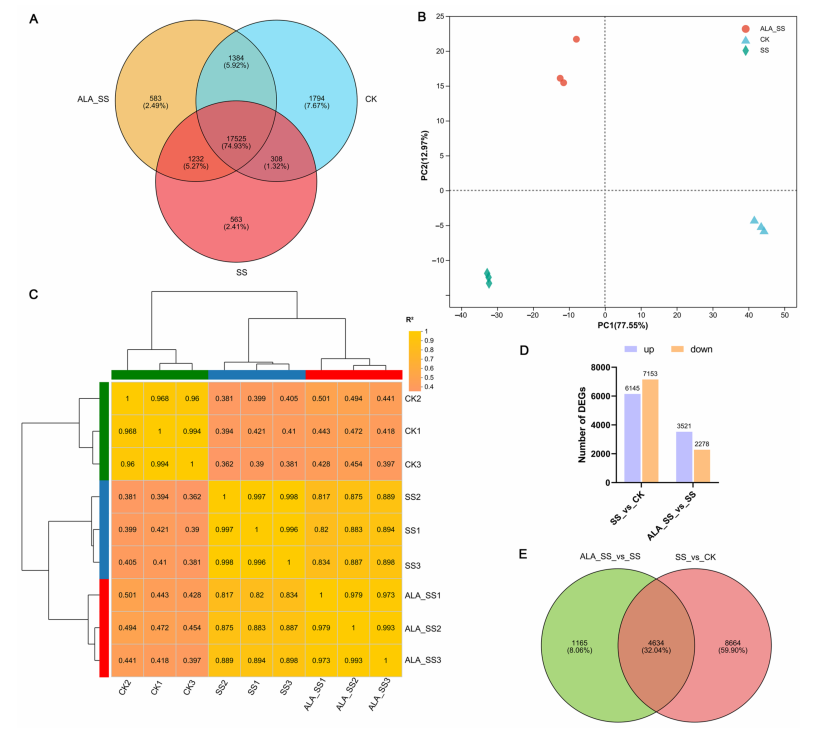
Figure 2. Analysis of transcriptome sequencing data.
Gene Ontology (GO) and KEGG enrichment analyses were conducted to investigate the functions of DEGs shared between the two comparison groups. GO classification revealed that in the Biological Process category, most DEGs were involved in “cellular processes,” “metabolic processes,” and “biological regulation.” In the Cellular Component category, DEGs were mainly associated with “cell part,” “organelle,” and “membrane part.” For Molecular Function, the dominant terms were “binding” and “catalytic activity.” These results indicate that ALA-regulated DEGs under salt stress are primarily involved in cellular activities, metabolism, and regulatory processes.
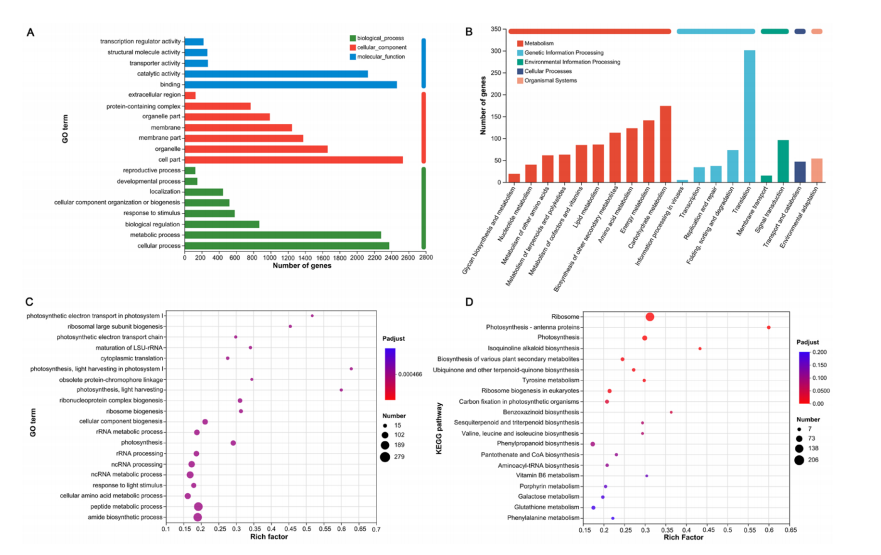
Figure 3. Functional annotation and enrichment analysis of shared DEGs conducted by GO and KEGG
Transcription factors (TFs) are key regulators in plant stress responses. In this study, DEGs under salt stress (SS_vs_CK) were associated with 45 TF families, mainly MYB-related, ERF, and MYB. Under ALA treatment (ALA_SS_vs_SS), 37 TF families were identified, with WRKY, NAC, and ERF being most common. Ten major TF families were shared between both comparisons—including NAC, MYB, WRKY, and bHLH—highlighting their central role in mediating the maize response to salt stress and ALA treatment.
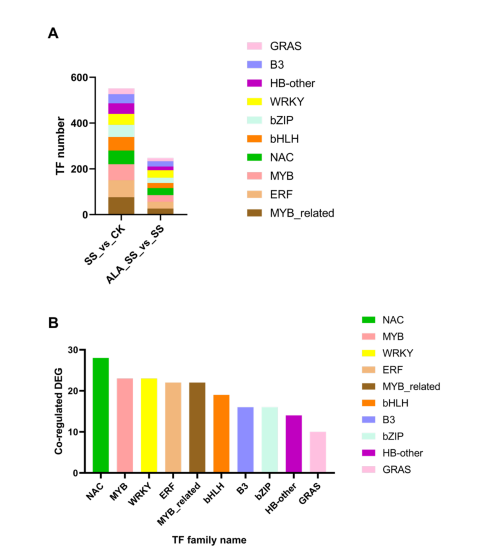
Figure 4. Analysis of transcription factors
Chlorophyll biosynthesis involves a series of enzymes catalyzed from L-glutamate. Under salt stress, several key genes involved in this pathway were down-regulated, indicating inhibited chlorophyll production. However, ALA treatment reversed this effect, up-regulating genes such as hemA, chlH, chlM, por, and chlG, suggesting that ALA promotes chlorophyll biosynthesis and may help maintain photosynthetic capacity in maize under salt stress.
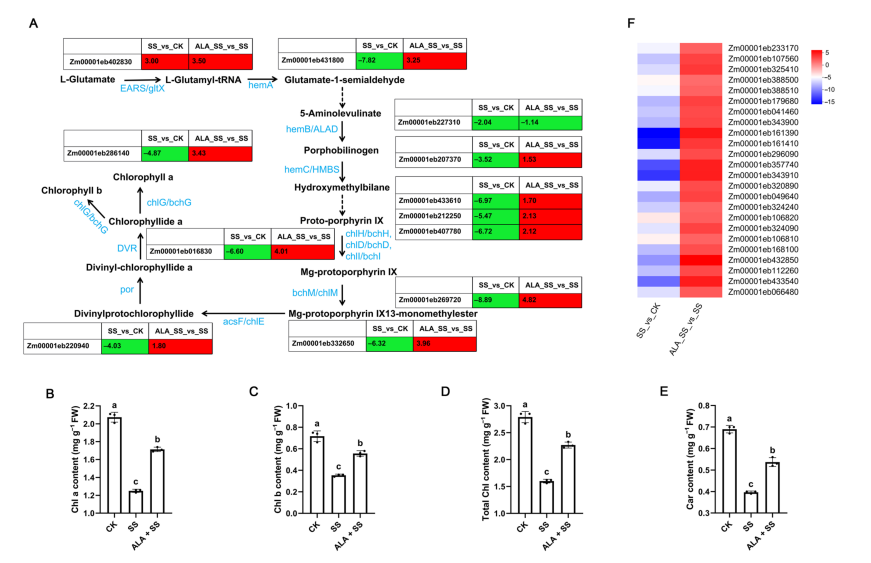
Figure 5. DEGs involved in porphyrin metabolism and photosynthesis-antenna proteins in two comparisons.
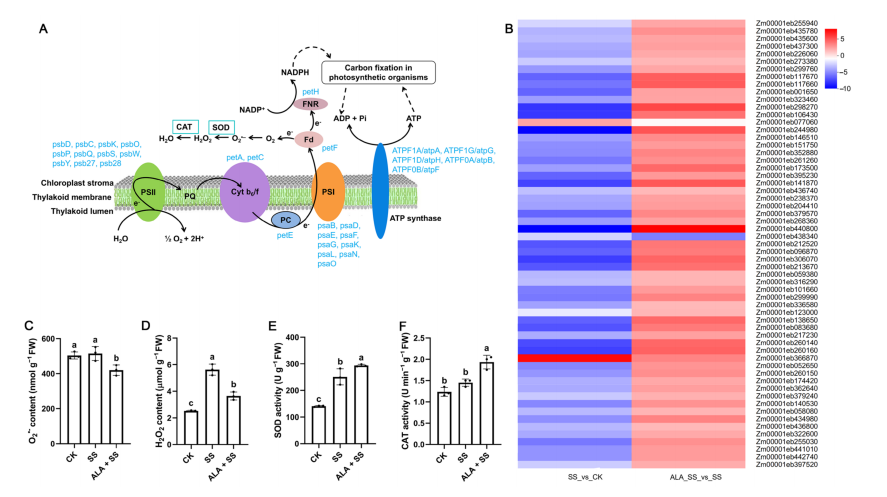
Figure 6. DEGs involved in photosynthesis in two comparisons.
The expression of key genes involved in the C4-dicarboxylic acid cycle and Calvin–Benson cycle showed significant changes under salt stress and ALA treatment. Important carbon fixation genes, including phosphoenolpyruvate carboxylase (ppc), malate dehydrogenase (maeB), and ribulose-bisphosphate carboxylase (rbcL and rbcS), were differentially expressed. Phosphoglycerate kinase (PGK) was down-regulated by salt stress but up-regulated with ALA supplementation. Other enzymes involved in carbon assimilation, such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH), triosephosphate isomerase (TPI), fructose-bisphosphate aldolase (ALDO), fructose-1,6-bisphosphatase (FBP), sedoheptulose-bisphosphatase, and phosphoribulokinase (PRK), also showed differential expression. These changes suggest that ALA helps maintain or enhance carbon fixation and sugar synthesis under salt stress.
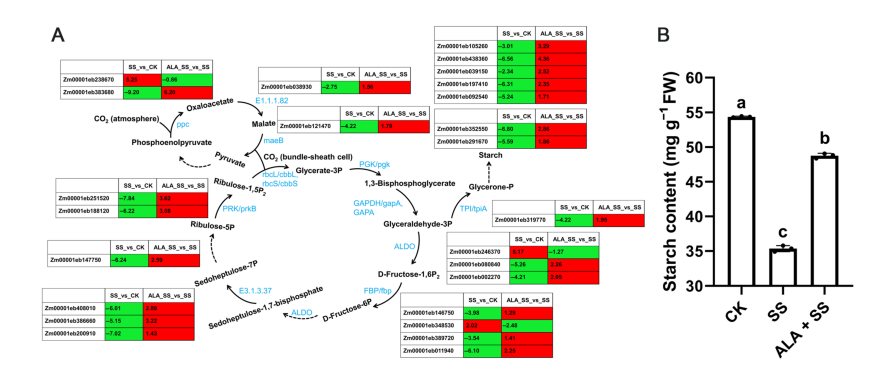
Figure 7. DEGs involved in carbon fixation in photosynthetic organisms in two comparisons
This study comprehensively analyzed transcriptomic changes in maize seedlings under salt stress with exogenous ALA treatment, revealing how ALA enhances salt tolerance. ALA improved photosynthesis, reducing and delaying salt-induced damage. Morphological observations showed ALA protected seedling growth and leaf cell structure. RNA-Seq identified 4634 shared DEGs, enriched in pathways related to porphyrin metabolism, photosynthesis, and carbon fixation. Physiological results demonstrated that ALA prevented pigment degradation, boosted antioxidant enzyme activities (SOD, CAT) to reduce ROS, and lessened starch content decline under salt stress. Key transcription factors, including NAC, MYB, and WRKY families, were involved in regulation. These findings highlight ALA’s role in protecting photosynthesis and stress tolerance, with potential benefits for maize growth, yield, and quality.
Related News
2025-06-04
Functional study of plant hormone transporters using a heterologous system
2025-05-29
2025-05-27
2025-05-22
2025-05-20
2025-05-16
2025-05-13
2025-05-09



