Literature Sharing | PbARP1 enhances salt tolerance of ‘Duli’ pear (Pyrus betulifolia Bunge) through abscisic acid signalling pathway
Release time:
2025-06-27
This study investigates salt stress tolerance in pears and identifies 35 salt-tolerant genes using a yeast expression library. Among them, PbARP1 was found to be significantly upregulated under salt stress in 'Duli' pear (Pyrus betulaefolia). Functional analyses showed that overexpression of PbARP1 in transgenic pear calli enhanced salt tolerance, while its suppression increased sensitivity to salt stress. Silencing PbARP1 also altered the expression of key ABA signaling genes, including PbPYL4, PbPYL9, PbPYL8, PbSRK2I, and PbABI5, suggesting that PbARP1 modulates salt stress responses through the ABA signaling pathway. Furthermore, PbPYL8, an ABA receptor, was identified as a protein interacting with PbARP1, highlighting its pivotal role in ABA-mediated salt stress regulation in pear.
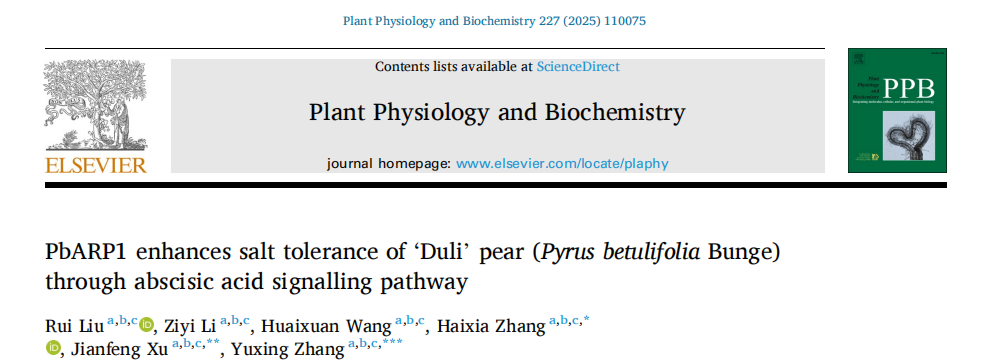
This study assessed the quality of RNA and cDNA for library construction. Agarose gel electrophoresis showed clear 28S and 18S rRNA bands, with a weak 5S band, and spectrophotometric analysis confirmed high RNA purity (OD260/280 between 1.8–2.2; OD260/230 >2), indicating intact, impurity-free RNA. Electrophoresis of synthesized ds cDNA showed diverse band sizes, suggesting good quality. After fragment homogenization and removal of small fragments, no bands below 500 bp were observed, confirming successful size selection. The constructed library, based on 688 monoclonal colonies, had a capacity of 6.88 × 10⁴ cfu/mL and a total of 1.376 × 10⁵ clones. PCR verification of 24 random colonies showed ~1000 bp inserts, indicating efficient fragment insertion and a high-quality cDNA library.
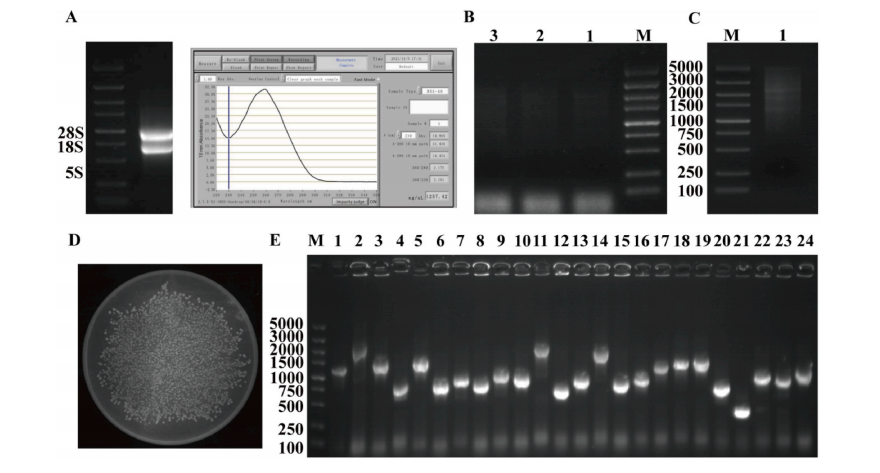
Fig. 1. Construction of yeast expression library and analysis of library quality in ‘Duli’ pear
This study evaluated salt tolerance by comparing yeast growth on SG-U medium with varying NaCl concentrations. At 1.5 M NaCl, the yeast library grew successfully, while the control did not, establishing 1.5 M NaCl as the screening condition. A total of 96 positive yeast clones were isolated, sequenced, and analyzed, resulting in 35 unique ‘Duli’ pear genes after removing redundant and empty sequences. Functional validation in yeast showed that all 35 clones grew on both SG-U and SG-U + 1.5 M NaCl plates, whereas the negative control only grew on SG-U. These results indicate that the 35 ‘Duli’ pear genes significantly enhanced yeast salt tolerance, suggesting their potential roles in salt stress resistance.
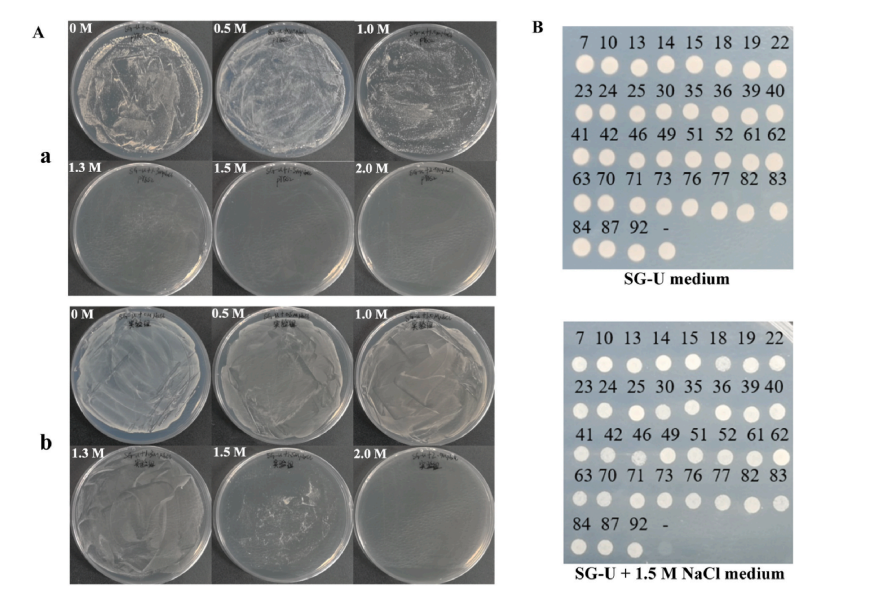
Fig. 2. Screening of candidate genes and verification analysis of yeast rotation
A total of 35 genes were identified from ‘Duli’ pear, representing diverse functional categories, including transcription factors, MAPK pathway-related kinases, disulfide isomerases, photosynthesis-related enzymes, and several proteins of unknown function. qPCR analysis under salt stress revealed that three genes (19, 25, and 62) were significantly upregulated over time, with gene 25—encoding auxin-repressed protein 1 (PbARP1)—showing the most pronounced response, peaking at 6 and 12 hours. Based on these results, PbARP1 was selected for further functional analysis to explore its regulatory role in salt stress tolerance.

Fig. 3. Heatmap of expression analysis of 35 genes in ‘Duli’ pear seedlings under salt stress at different time periods.
To assess the role of PbARP1 in salt stress response, transgenic pear calli overexpressing PbARP1 were generated using a pCAMBIA2301 vector under the 35S promoter. Successful transformation was confirmed by GUS staining, PCR, and qRT-PCR in two lines (L1 and L2). Under normal conditions, transgenic and wild-type (WT) calli showed no visible differences. However, after salt stress treatment, transgenic calli exhibited improved growth, higher fresh weight and proline content, and lower levels of relative electric conductivity (REC), hydrogen peroxide (H₂O₂), and malondialdehyde (MDA), compared to WT. DAB and NBT staining also showed reduced reactive oxygen species (ROS) accumulation in transgenic lines. These physiological and phenotypic results indicate that PbARP1 overexpression enhances salt tolerance in pear calli.
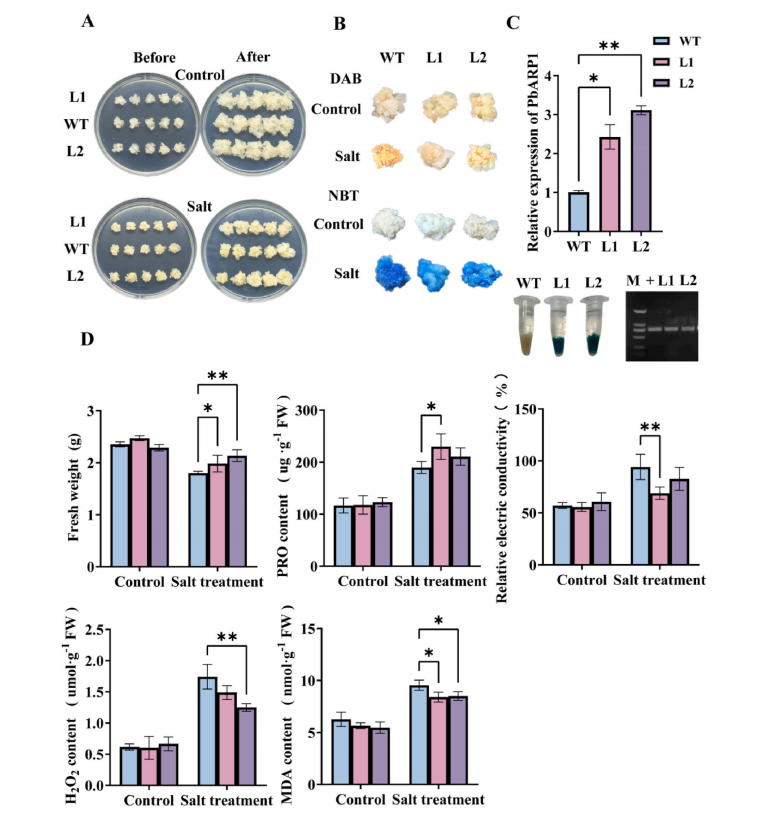
Fig. 4. Analyses of phenotype and physiological indexes of PbARP1-overexpressing transgenic pear calli under salt stress
To investigate the function of PbARP1 in salt tolerance, a VIGS (virus-induced gene silencing) approach was used to silence PbARP1 in pear plants. Transcript levels of PbARP1 were reduced by 62% in silenced plants (PbARP1-TRV2) compared to the empty vector control (EV). Under normal conditions, PbARP1-TRV2 and EV plants showed no noticeable differences. However, under 200 mM NaCl stress, PbARP1-TRV2 plants exhibited more severe wilting and necrosis. Chlorophyll content was significantly lower in silenced plants under salt stress, and histochemical staining revealed greater ROS accumulation in PbARP1-TRV2 plants. Physiological measurements showed no significant differences under control conditions, but under salt stress, silenced plants had lower fresh weight and proline levels, and higher MDA and H₂O₂ levels than controls. These results indicate that PbARP1 plays a positive role in enhancing salt tolerance in pear.
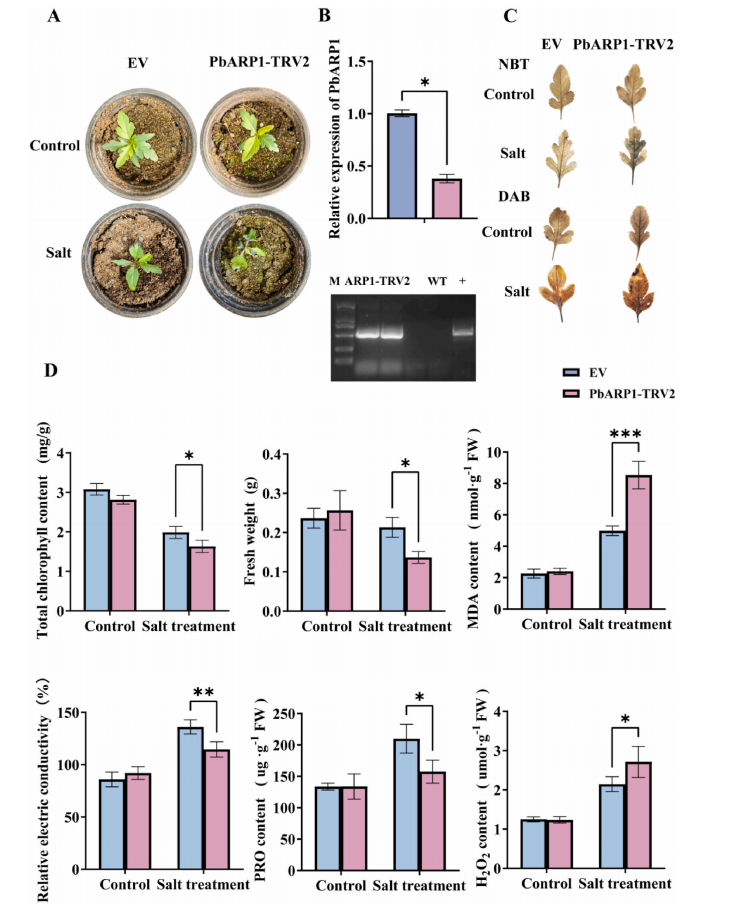
Fig. 5. Analyses of phenotype and physiological indexes of PbARP1 silenced ‘Duli’ seedlings under salt stres
To explore the regulatory role of PbARP1 in the ABA signaling pathway under salt stress, ABA levels and the expression of ABA-related genes were analyzed in transgenic and PbARP1-silenced ‘Duli’ pear plants. Under normal conditions, ABA levels were similar between WT and PbARP1-overexpressing calli, but salt stress induced higher ABA accumulation in WT than in transgenic lines. Despite lower ABA levels, transgenic calli showed significantly upregulated expression of ABA signaling genes, especially PbPYL8 and PbPYL9, under salt stress. In contrast, PbARP1-silenced plants (PbARP1-TRV2) exhibited reduced ABA levels and markedly decreased expression of PbPYL4, PbPYL8, PbPYL9, PbSRK2I, and PbABI5 under salt stress, with PbPYL8 and PbSRK2I being most affected. These results suggest that PbARP1 enhances salt tolerance by modulating the ABA signaling pathway.
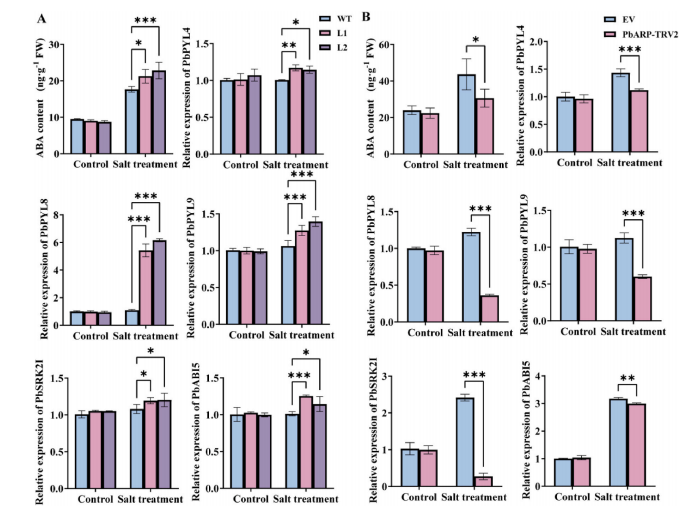
Fig. 6. ABA content and relative expression levels of ABA-related genes in transgenic pear calli (A) and silenced ‘Duli’ pear plants (B).
To elucidate how PbARP1 functions in ABA signaling, yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays were conducted. The results demonstrated that PbARP1 specifically interacts with the ABA receptor PbPYL8, but not with PbPYL4, PbPYL9, PbSRK2I, or PbABI5. BiFC assays confirmed this interaction in vivo. Moreover, ABA treatment had no significant effect on the interaction between PbARP1 and PbPYL8 in the Y2H system. These findings indicate that PbARP1 physically interacts with PbPYL8, suggesting that PbARP1 is a key component of the ABA core signaling pathway in pear.
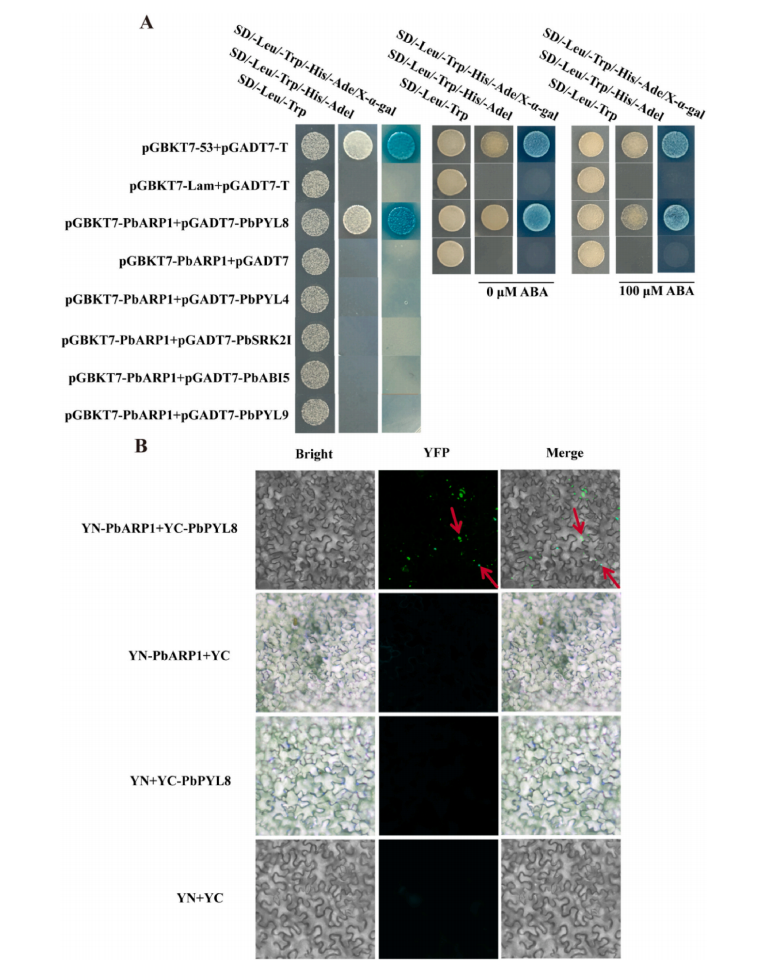
Fig. 7. Screening and verification of PbARP1’s interacting proteins by Y2H (A) and BiFC (B) assays.
This study identified 35 salt-tolerant genes in pears through yeast cDNA library screening and explored the role of PbARP1 in salt tolerance. A high-quality cDNA library was constructed, revealing involvement of pathways such as MAPK-related serine/threonine kinases, disulfide isomerase, and photosynthetic enzymes in salt stress response. Overexpression of PbARP1 enhanced salt tolerance in transgenic pears, while silencing PbARP1 increased sensitivity. PbARP1 improved salt tolerance by upregulating salt-responsive genes (PbPYL4, PbPYL9, PbPYL8, PbSRK2I, and PbABI5), and its interaction with the ABA receptor PbPYL8 was confirmed. These findings provide a basis for understanding the molecular mechanisms of ARP1-mediated salt tolerance in pears.
Related News
2025-06-27
2025-06-24
2025-06-20
2025-06-17
2025-06-12
2025-06-10
2025-06-06
Literature Sharing | SPL1 positively regulates cuticular ridge wax biosynthesis in Arabidopsis
2025-06-04
Functional study of plant hormone transporters using a heterologous system



