Literature Sharing | Phosphorylation of the strawberry MADS-box CMB1 regulates ripening via the catabolism of abscisic acid
Release time:
2025-06-24
This study uncovers a regulatory mechanism linking transcriptional control and posttranslational modification in strawberry fruit ripening. The MADS-box transcription factor FaCMB1 acts as a negative regulator of ripening, with both its transcript and protein levels decreasing during fruit development, a process enhanced by ABA. Functional manipulation of FaCMB1 significantly affected ripening and ABA content.
RNA-seq and promoter assays revealed that FaCMB1 represses FaASR, which in turn inhibits FaCYP707A4, a gene involved in ABA catabolism. Notably, FaCMB1 undergoes phosphorylation by the kinase FaSTPK, and this posttranslational modification increases during ripening, weakening FaCMB1’s repressive effect on FaASR.
The authors propose a feedback loop: rising ABA levels reduce FaCMB1 expression and increase its phosphorylation, lifting repression on FaASR, which further suppresses FaCYP707A4, leading to ABA accumulation and promotion of fruit ripening. This work highlights the role of protein phosphorylation in fine-tuning transcriptional regulation during fleshy fruit ripening.

To identify MADS-box genes involved in octoploid strawberry fruit ripening, a spatiotemporal transcriptome analysis across three developmental stages (green, turning, and half red) was conducted. Six candidate genes were identified based on their expression patterns, domain structure, and evolutionary relationships with Arabidopsis homologs, including FaCMB1, an ortholog of AtSEP4.
FaCMB1 was primarily expressed in reproductive organs and showed a progressive decline in both transcript and protein levels during fruit development. Subcellular localization confirmed its presence in the nucleus. qPCR and immunoblotting analyses validated these patterns, and exogenous ABA treatment further suppressed FaCMB1 expression, both at the transcript and protein levels. The presence of ABA-responsive elements in the FaCMB1 promoter supports its ABA-mediated regulation.
These results collectively suggest that FaCMB1 functions as a negative regulator of strawberry fruit ripening and is responsive to hormonal cues such as ABA.
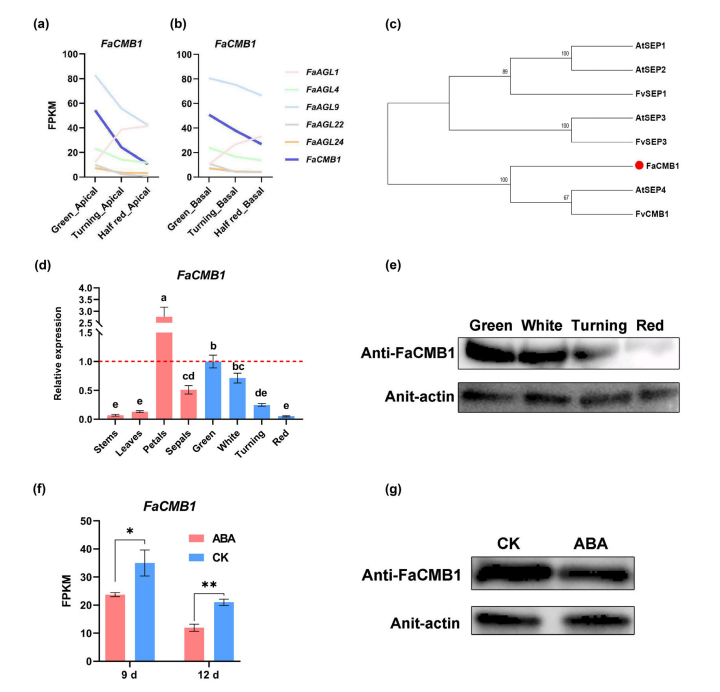
Fig. 1 The expression level of FaCMB1 is closely correlated with the ripening process and responds to abscisic acid (ABA).
To investigate the function of FaCMB1 in strawberry fruit ripening, a reliable transient expression system was used to overexpress or silence FaCMB1 in green-stage octoploid ‘Yuexin’ strawberries. The results showed that silencing FaCMB1 accelerated ripening, while overexpression delayed it, confirming its role as a ripening repressor.
Corresponding changes in FaCMB1 transcript and protein levels were observed. HPLC analysis revealed that ABA levels increased by 1.5-fold in FaCMB1-RNAi fruit and decreased by approximately 30% in FaCMB1-OE fruit compared to controls. GC-MS analysis showed that sugars (fructose, glucose, sucrose) significantly increased and citric acid decreased in FaCMB1-RNAi fruit, whereas FaCMB1 overexpression inhibited sugar accumulation and delayed citric acid degradation.
Additionally, the furanone content, a key aroma compound, was elevated by approximately 1.3-fold in FaCMB1-silenced fruit and reduced by about 66% in overexpressed samples. These findings demonstrate that FaCMB1 negatively regulates strawberry ripening by modulating ABA levels and ripening-related metabolic traits.
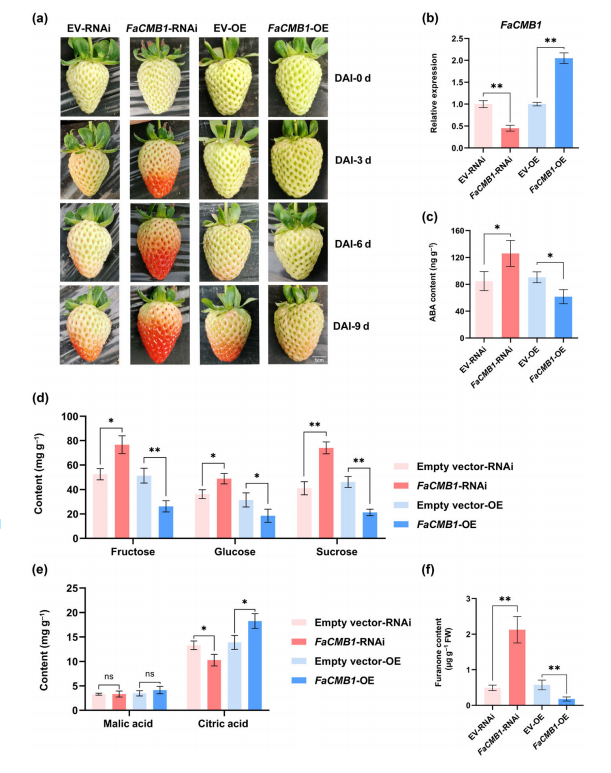
Fig. 2 FaCMB1 represses octoploid strawberry fruit ripening and affects the development of quality-related traits
To identify cofactors of FaCMB1, a cDNA library from octoploid strawberry fruit was screened using FaCMB1 as bait, leading to the identification of 64 positive clones. Among them, FaMYB10, a key regulator of anthocyanin biosynthesis, was identified as a potential interactor. The interaction between FaCMB1 and FaMYB10 was validated through yeast two-hybrid (Y2H), bimolecular fluorescence complementation (BiFC), and luciferase complementation imaging (LCI) assays.
Although FaMYB10 expression remained unchanged in FaCMB1 overexpression or silencing lines, and its promoter was not directly regulated by FaCMB1, the expression of downstream anthocyanin biosynthesis genes was significantly affected. Specifically, these genes were activated in FaCMB1-RNAi fruit and repressed in FaCMB1-OE fruit. Consistently, anthocyanin accumulation increased upon FaCMB1 silencing and decreased with its overexpression.
These results suggest that FaCMB1 may repress anthocyanin biosynthesis by interacting with FaMYB10 at the protein level rather than through transcriptional regulation.
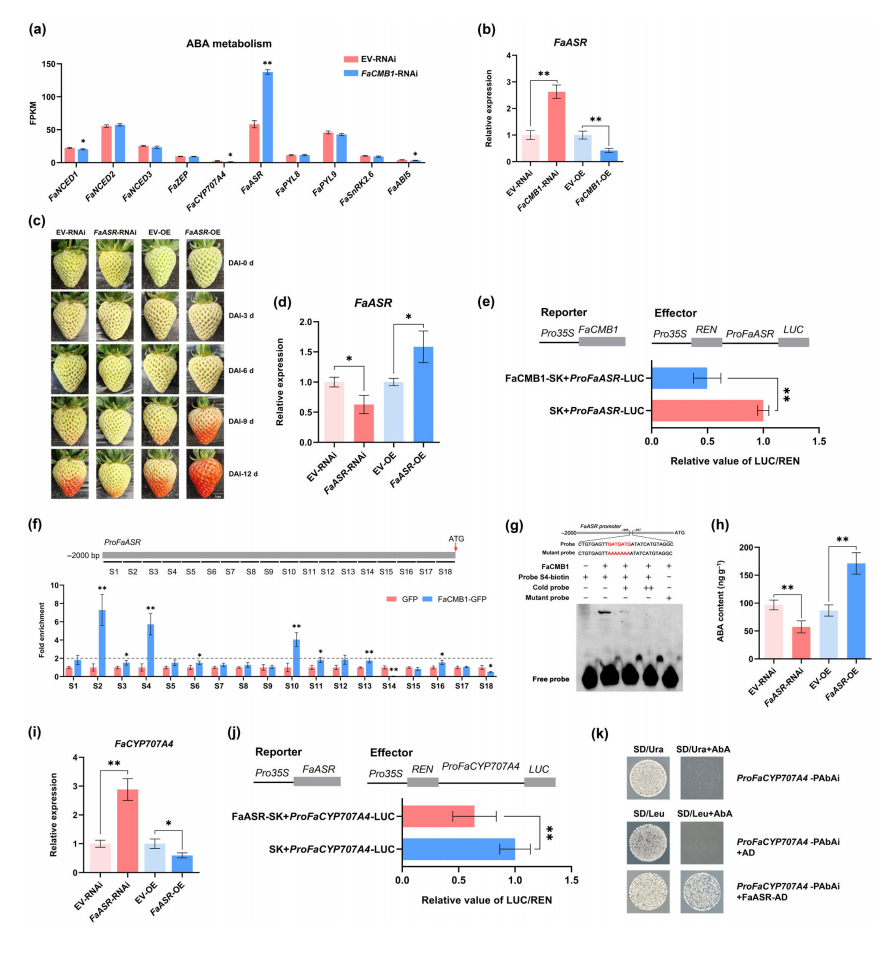
Fig. 3 Transcriptional cascade of FaCMB1-FaASR-FaCYP707A module controls abscisic acid (ABA) catabolism and thereby regulates fruit ripening.
To identify cofactors of FaCMB1, a cDNA library from octoploid strawberry fruit was screened using FaCMB1 as bait, leading to the identification of 64 positive clones. Among them, FaMYB10, a key regulator of anthocyanin biosynthesis, was identified as a potential interactor. The interaction between FaCMB1 and FaMYB10 was validated through yeast two-hybrid (Y2H), bimolecular fluorescence complementation (BiFC), and luciferase complementation imaging (LCI) assays.
Although FaMYB10 expression remained unchanged in FaCMB1 overexpression or silencing lines, and its promoter was not directly regulated by FaCMB1, the expression of downstream anthocyanin biosynthesis genes was significantly affected. Specifically, these genes were activated in FaCMB1-RNAi fruit and repressed in FaCMB1-OE fruit. Consistently, anthocyanin accumulation increased upon FaCMB1 silencing and decreased with its overexpression.
These results suggest that FaCMB1 may repress anthocyanin biosynthesis by interacting with FaMYB10 at the protein level rather than through transcriptional regulation.
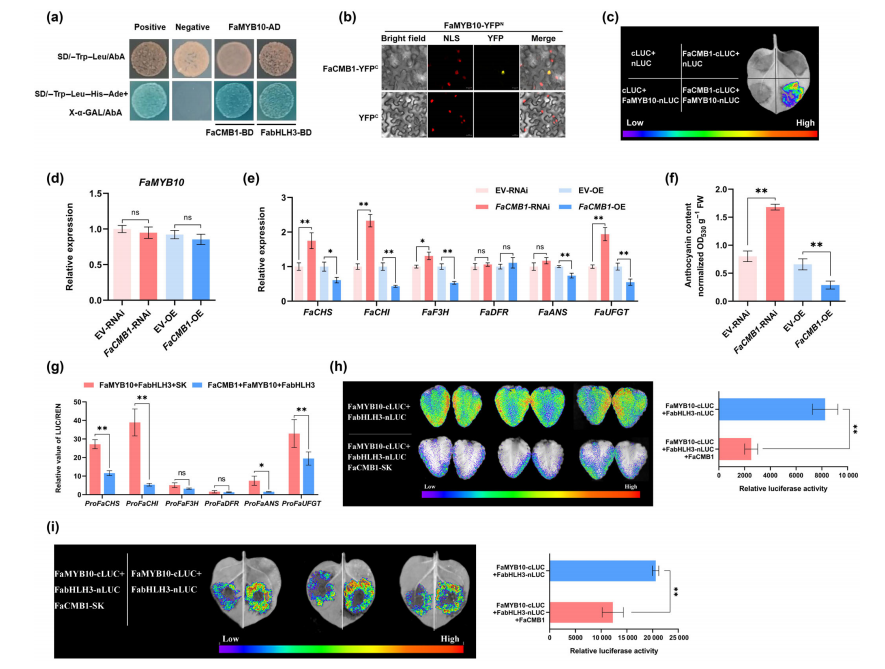
Fig. 4 FaCMB1 interacts with FaMYB10 and attenuates the transcriptional activation of anthocyanin biosynthesis genes by FaMYB10-FabHLH3.
Phosphorylation is a key regulatory mechanism for transcription factor activity. Through yeast two-hybrid (Y2H) library screening, FaSTPK (Isoform0051676) was identified as an interactor of FaCMB1. Its kinase activity was confirmed by an in vitro kinase assay. The interaction between FaCMB1 and FaSTPK was further validated in vivo using BiFC and LCI assays, and confirmed in vitro through GST pull-down assays. These results suggest that FaSTPK physically interacts with FaCMB1 and may phosphorylate it to regulate its function.
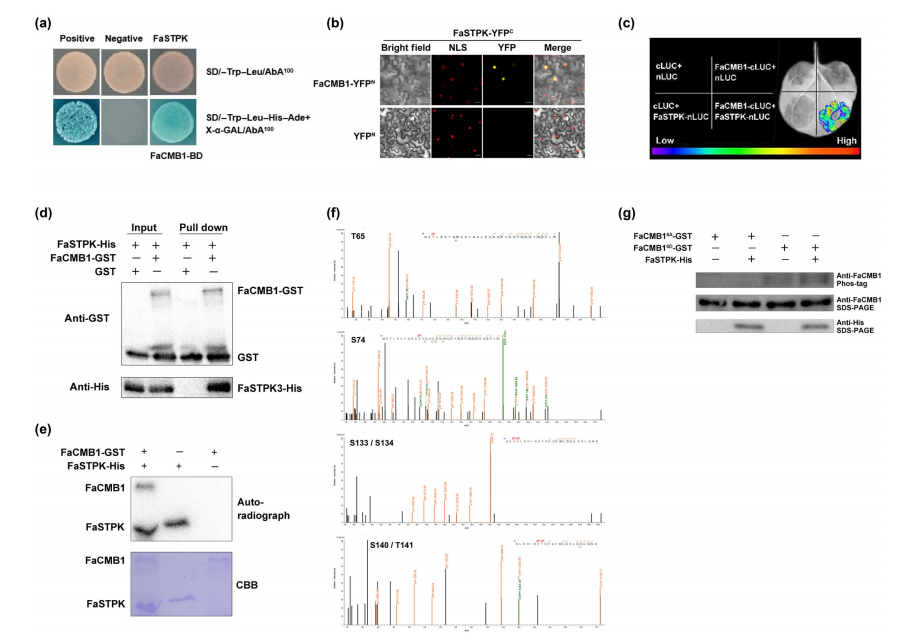
Fig. 5 FaCMB1 physically interacts with and is phosphorylated by FaSTPK at specific sites.
To assess the functional impact of FaSTPK-mediated phosphorylation on FaCMB1, dual luciferase assays were performed to compare the transcriptional activity of different FaCMB1 phosphorylation variants. Both wild-type FaCMB1 and the non-phosphorylatable mutant FaCMB16A strongly repressed FaASR transcription, while the phospho-mimic mutant FaCMB16D showed a significantly reduced repression effect.
ChIP-qPCR assays further revealed that FaCMB16D-GFP had about 60% lower enrichment at the S4 region of the FaASR promoter compared to FaCMB1-GFP, indicating weakened DNA binding. In contrast, FaCMB16A-GFP showed no significant difference in promoter binding. These results demonstrate that phosphorylation of FaCMB1 reduces its ability to bind to the FaASR promoter and weakens its transcriptional repression function.
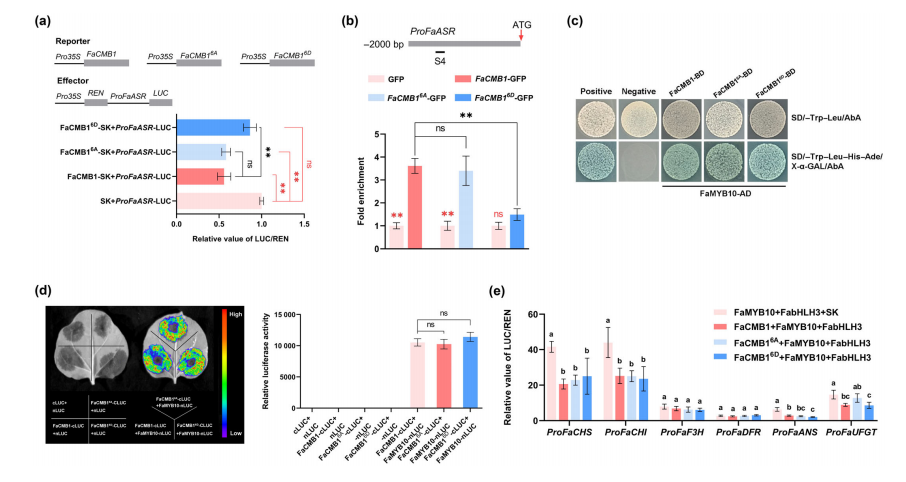
Fig. 6 FaSTPK-mediated phosphorylation alleviates the transcriptional repression of FaCMB1.
This study investigated the role of FaSTPK-mediated phosphorylation of FaCMB1 during strawberry fruit ripening. Real-time PCR showed that FaSTPK expression peaked at the turning stage and was unaffected by ABA. Immunoblotting revealed that FaCMB1 phosphorylation increased during ripening, peaking at the turning stage, matching FaSTPK expression patterns.
Transient overexpression of wild-type FaCMB1 and the non-phosphorylatable mutant FaCMB16A significantly delayed fruit ripening, while the phospho-mimic FaCMB16D showed no inhibitory effect. Transcript and protein levels of FaCMB1 were elevated in all transgenic lines. Phos-tag assays confirmed decreased phosphorylation in FaCMB16A-OE fruit and increased phosphorylation in FaCMB16D-OE fruit.
Overexpression of FaCMB1 and FaCMB16A reduced FaASR transcript levels and increased FaCYP707A4 expression, whereas FaCMB16D overexpression had no effect, resulting in higher ABA accumulation. Correspondingly, anthocyanin and furanone levels mirrored ABA content changes.
These results demonstrate that FaCMB1 phosphorylation modulates its transcriptional repression activity and plays a critical role in regulating strawberry fruit ripening.
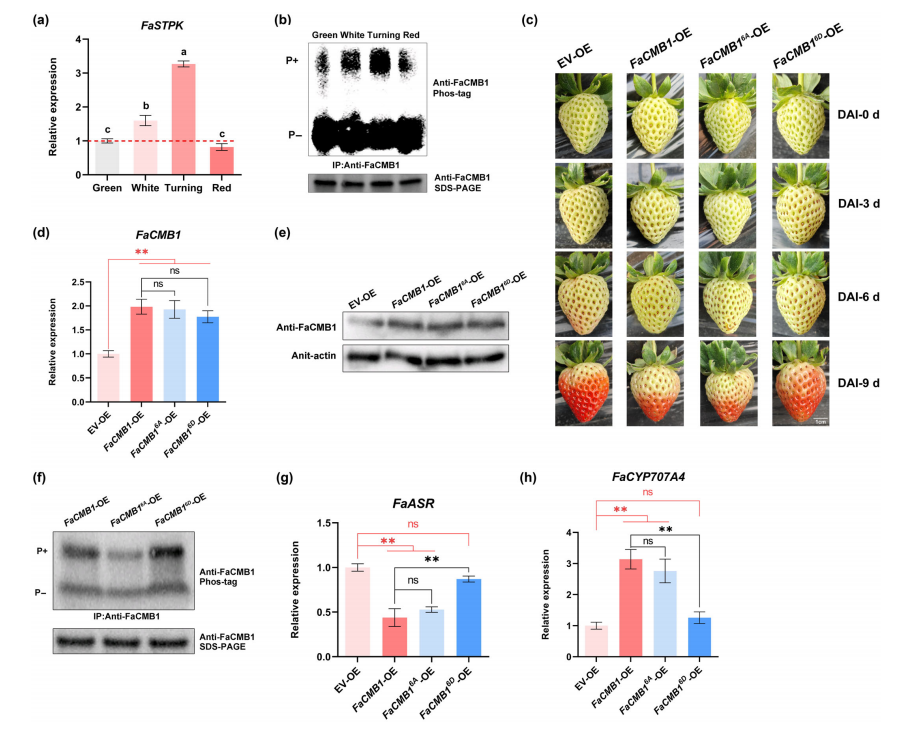
Fig. 7 The phosphorylation of FaCMB1 is involved in the control of strawberry fruit ripening.
In summary, the study proposes a model in which FaCMB1 acts as a negative regulator of strawberry fruit ripening by repressing FaASR, an activator that inhibits ABA catabolism via FaCYP707A4. During ripening, rising ABA levels reduce FaCMB1 transcript and protein abundance, lessening its repression of FaASR. Simultaneously, increased phosphorylation of FaCMB1 diminishes its DNA-binding ability, further relieving repression on FaASR. This leads to enhanced suppression of FaCYP707A4, promoting ABA accumulation and fruit ripening. Additionally, reduced FaCMB1 weakens its interaction with FaMYB10, allowing FaMYB10 to partner with FabHLH3 and activate anthocyanin biosynthesis. The study reveals how FaCMB1 integrates transcriptional and posttranslational regulation to finely control strawberry fruit ripening.
Related News
2025-06-24
2025-06-20
2025-06-17
2025-06-12
2025-06-10
2025-06-06
Literature Sharing | SPL1 positively regulates cuticular ridge wax biosynthesis in Arabidopsis
2025-06-04
Functional study of plant hormone transporters using a heterologous system
2025-05-29



