Literature Sharing | Overexpression of soybean flavonoid 3′-hydroxylase enhances plant salt tolerance by promoting ascorbic acid biosynthesis
Release time:
2025-06-20
This study reveals that the flavonoid 3′-hydroxylase gene GmF3′H plays a key role in enhancing salt tolerance in soybean by regulating redox homeostasis. Using CRISPR/Cas9 knockout and overexpression approaches, the researchers demonstrated that GmF3′H competitively interacts with CSN5B, disrupting its binding to VTC1, a key enzyme in ascorbic acid biosynthesis. This redirection of metabolic flux toward the L-galactose pathway leads to increased ascorbic acid (AsA) levels, enhancing ROS scavenging capacity and improving salt stress tolerance during seed germination and seedling growth.

GmF3′H (Glyma.06G202300) encodes a 513-amino acid protein and is highly conserved among plant species, showing clear divergence between monocots and dicots. RNA-seq analysis revealed that genes upstream of GmF3′H in the flavonoid biosynthesis pathway are predominantly expressed in the seed coat of soybeans. To explore GmF3′H’s role in salt stress response, researchers used a natural mutant with a non-functional GmF3′H gene (bsc) and generated both overexpression lines (GmF3′H-OE) and CRISPR/Cas9 knockout lines (gmf3′h) in soybean. Salt tolerance assays showed that GmF3′H overexpression significantly improved seed germination under salt stress, while knockout lines were more sensitive. GmF3′H also increased flavonoid accumulation and anthocyanin pigmentation in the seed coat, enhancing antioxidant capacity and contributing to improved salt tolerance.
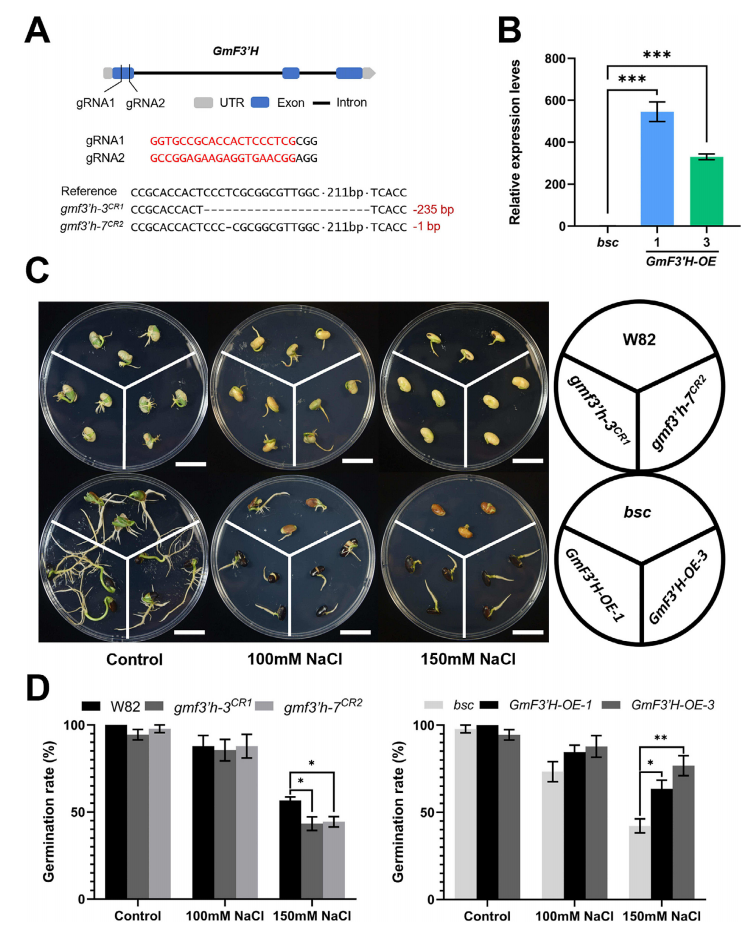
Fig. 1. GmF3’H confers salt tolerance during soybean seeds germination.
During salt stress at the seedling stage, GmF3′H-overexpressing (OE) soybean lines showed significantly higher root density and survival rates compared to the bsc mutant. GmF3′H-OE seedlings had reduced levels of MDA and H₂O₂, indicating enhanced ROS scavenging capacity. However, no notable differences in pigment accumulation or flavonoid content were observed between OE lines and bsc, suggesting that the improved salt tolerance at the seedling stage is independent of flavonoid compounds. These results imply that GmF3′H enhances antioxidant capacity and salt stress tolerance through flavonoid-independent molecular mechanisms.
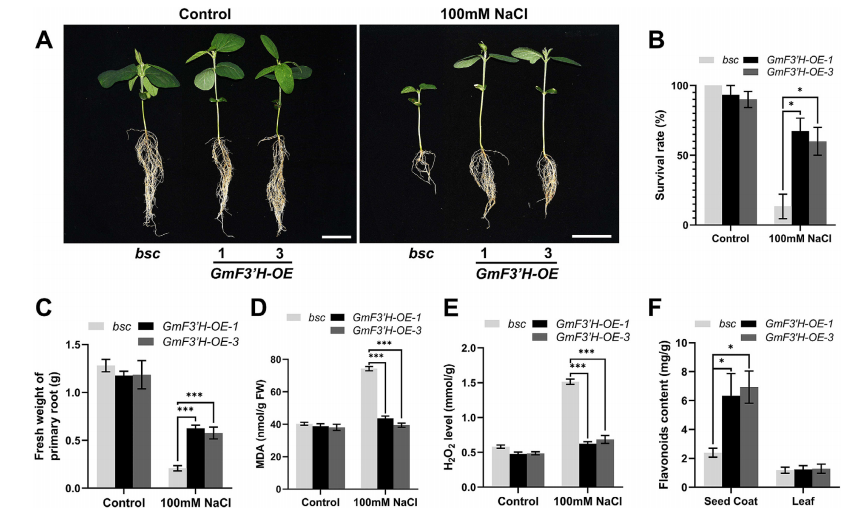
Fig. 2. Overexpressing GmF3’H enhances seedlings’ salt tolerance in soybean.
To investigate how GmF3′H protects soybean seedlings from salt stress, subcellular localization analysis revealed that GmF3′H is localized in the endoplasmic reticulum (ER). Yeast two-hybrid screening identified CSN5B as a strong interacting partner of GmF3′H, and this interaction was further confirmed by luciferase complementation assay, supporting a potential functional link between GmF3′H and CSN5B in stress response.
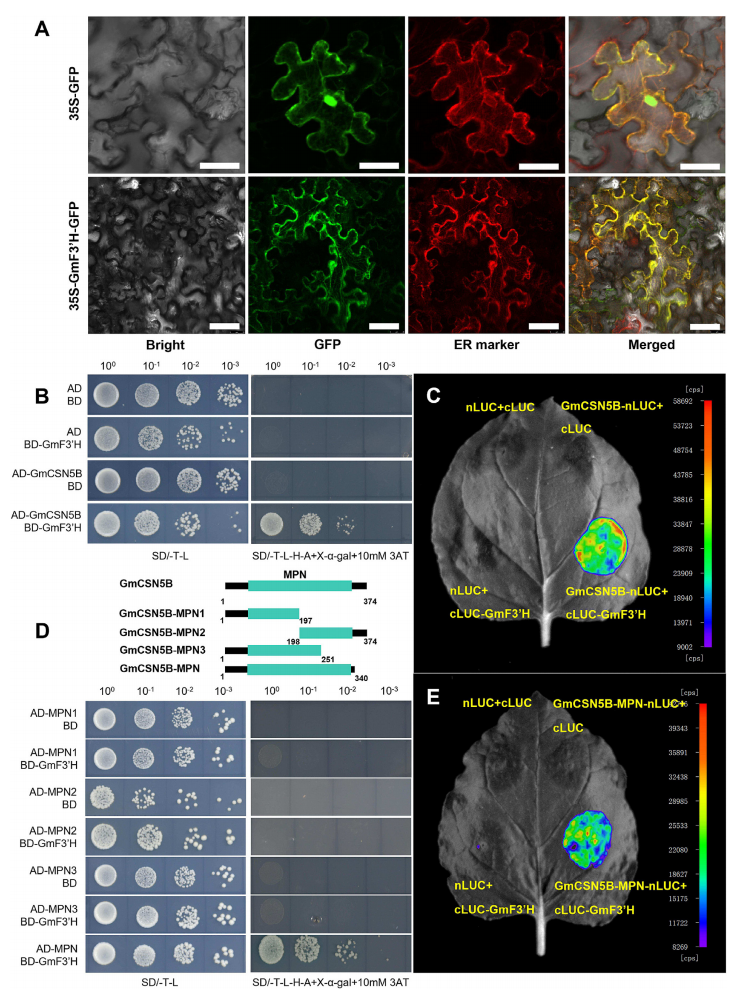
Fig. 3. GmF3’H interacts with GmCSN5B.
Protein interaction and localization analyses revealed that GmF3′H, CSN5B, and VTC1 co-localize in the endoplasmic reticulum, enabling competitive binding. Luciferase complementation assays confirmed that GmF3′H competes with VTC1 for binding to CSN5B in both soybean and Arabidopsis. Increasing GmF3′H levels reduced the interaction signal between CSN5B and VTC1, while excess VTC1 could restore the interaction, indicating competitive displacement. This competition likely prevents CSN5B-mediated degradation of VTC1, thereby promoting ascorbic acid (AsA) biosynthesis. Supporting this, AsA levels were significantly higher in GmF3′H-overexpressing lines and lower in gmf3′h mutants, particularly in tissues with high GmF3′H expression, confirming GmF3′H's role in enhancing salt tolerance by boosting AsA accumulation.
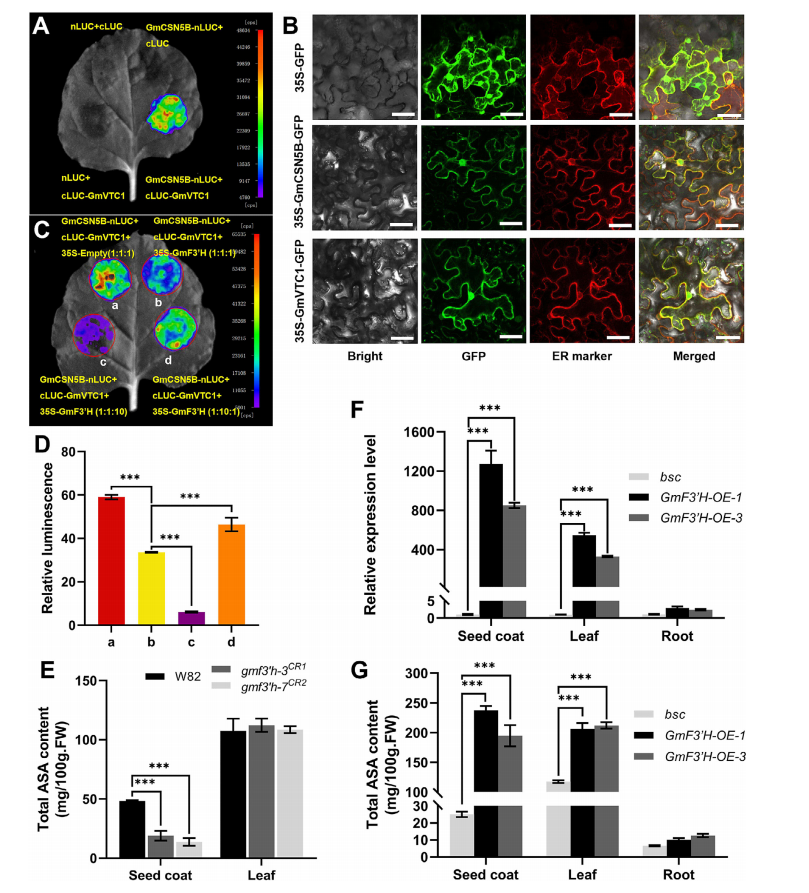
Fig. 4. GmF3’H increases ASA content in soybean tissues by inhibiting the degradation of GmVTC1 by GmCSN5B
The salt tolerance of the tt7 mutant (lacking F3′H) and wild-type Col-0 in Arabidopsis thaliana was evaluated. Results showed that Col-0 seeds exhibited greater salt tolerance compared to tt7 seeds (Fig. 5), suggesting that the AtF3′H gene, an ortholog of GmF3′H, also plays a significant role in enhancing salt tolerance in Arabidopsis thaliana seeds.
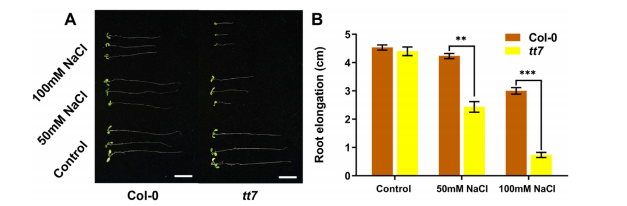
Fig. 5. AtF3’H gene confers seeds’ salt tolerance in Arabidopsis thaliana
Three stable Arabidopsis lines overexpressing GmF3′H were generated, showing significantly increased ascorbic acid (AsA) levels compared to wild-type Col-0. Correspondingly, these overexpression lines exhibited enhanced salt tolerance, indicating that the conserved F3′H gene improves plant salt stress resistance by boosting AsA-mediated reactive oxygen species (ROS) scavenging.
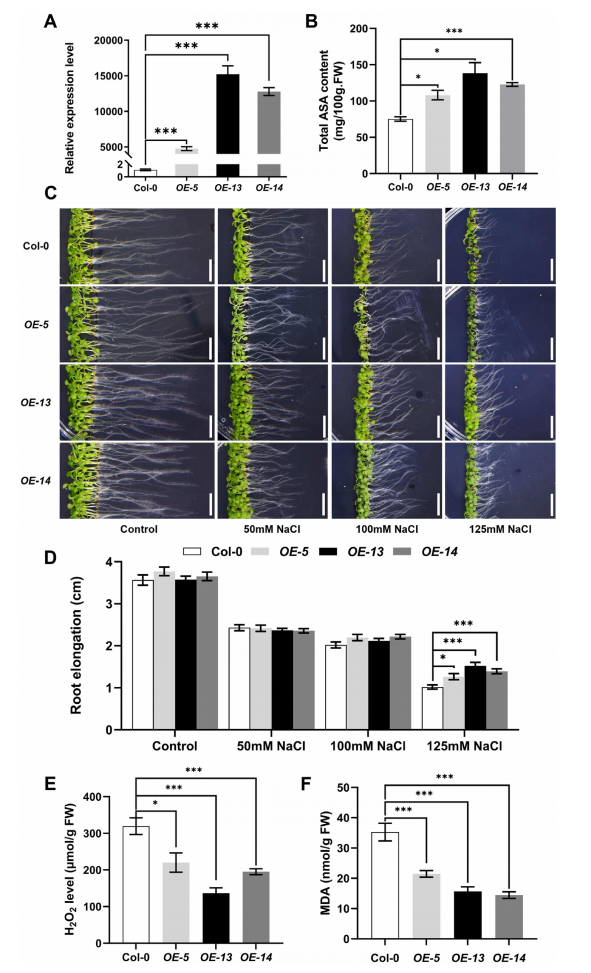
Fig. 6. Overexpression of GmF3’H improves seedling salt tolerance in Arabidopsis thaliana.
The identification of F3′H’s role in salt tolerance offers a valuable foundation for crop genetic improvement. This study advances understanding of plant salt stress mechanisms and presents novel approaches to enhance broad-spectrum salt tolerance, which is vital for crop food security.
Related News
2025-06-20
2025-06-17
2025-06-12
2025-06-10
2025-06-06
Literature Sharing | SPL1 positively regulates cuticular ridge wax biosynthesis in Arabidopsis
2025-06-04
Functional study of plant hormone transporters using a heterologous system
2025-05-29
2025-05-27



